The hydrophobic effect is considered to be the major driving force for the folding of globular proteins. It results in the burial of the hydrophobic residues in the core of the protein. It is exemplified by the fact that oil and water do not mix and was described well by G. S. Hartley in 1936 .
"The antipathy of the paraffin chain for water is, however, frequently misunderstood. There is no question of actual repulsion between individual water molecules and paraffin chains, nor is there any very strong attraction of paraffin chains for one another. There is, however, a very strong attraction of water molecules for one another in comparison with which the paraffin-paraffin or paraffin-water attractions are slight."
The thermodynamic factors which give rise to the hydrophobic effect are
complex and still incompletely understood. The free energy of transfer of
a non-polar compound from some reference state, such as an organic solution,
into water, Gtr, is made up of an enthalpy,
H, and entropy, -T
S, term.
At room temperature, the enthalpy of transfer from organic solution into aqueous solution is negligible; the interaction enthalpies are the same in both cases.
The entropy however is negative. Water tends to form ordered cages around the non-polar molecule and this leads to a decrease in entropy. At high temperatures (~ 110°C) these cages are no longer any stronger than bulk water, and the entropy contribution tends to zero. The enthalpy of transfer, however, is now positive (unfavourable). Because the temperature dependence of entropy and enthalpy are not the same, there is some temperature at which the hydrophobic effect is strongest, and the effect decreases at temperatures above and below this temperature. The decrease in the strength of the hydrophobic effect with decreasing temperatures is probably the major cause of cold-denaturation in proteins.
The contribution of the hydrophobic effect to globular protein stability has been estimated empirically both by measuring the thermodynamics of transfer of model compounds (e.g. blocked amino acids, cyclic peptides...) from organic solvents to water, and by site directed mutagenesis studies on proteins. The number arrived at is usually given as a function of the change in the solvent accessible non-polar surface area upon going from the unfolded to the folded state.
The model compound studies predict that the hydrophobic effect of exposing one buried methylene group to bulk water is 0.8 kcal/mol (in Pace, 1995). The site directed mutagenesis studies yielded a larger number with greater statistical variation: the average hydrophobic effect estimated by SDM for a buried methylene group is about 1.3 kcal/mol. However, when the SDM results for methylene were plotted against the size of the cavity created by the residue substitution, and extrapolated to zero, the result at zero cavity size is 0.8 kcal/mol - in agreement with the value found for the transfer of model compounds from octanol to water (Pace et al., 1996 and references therein). In the SDM studies, cavities created by residue substitution have an additional destabilizing effect: the loss of favourable VDWs interactions (as compared to the wild-type). Thus, the "hydrophobic effect" measured by SDM includes both an entropic component due to solvent ordering and a (primarily) enthalpic component due to loss of VDWs contacts within the protein.
Such an SDM study of T4 lysozyme replaced the 80% buried Ile3 residue
by Val (Eriksson et al,
1992): the loss of this methyl group gave rise to a decrease in stability
of 0.6 kcal/mol (corrected to 100% burial). This is smaller than expected
(c.f. 0.8 kcal/mol for methylene) and suggests that the mutation introduced
some smaller stabilizing influence, perhaps such as the alleviation of strain
within the protein.
 Click here for a gif showing
the cavity and links to the structures.
Click here for a gif showing
the cavity and links to the structures.
In barnase, 15 mutants were constructed in which a hydrophobic interaction
was deleted (V10A, V36A, V45A, I4A, I25A, I51A, I55A, I76A, I109A, I4V,
I25V, I51V, I55V, I76V & I109V). The finding was a strong correlation
between the degree of destabilization (which ranges from 0.60 to 4.71 kcal/mol)
and the number of methyl or methylene side chain groups surrounding the
methyl or methylene group that was deleted (r = 0.91) (Serrano
et al, 1992). See figure below.
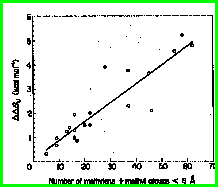
The average free energy decrease for removal of a completely buried methylene group was found to be 1.5±0.6 kcal/mol. This is additive, such that Ile or Leu to Ala can destabilize a protein by up to 5 kcal/mol. (Remember that many mesophilic proteins are stable by <10 kcal/mol, so two deletions such as this would be enough to destabilize a protein completely).
The figure below shows the backbone of barnase with the hydrophobic side-chains coloured blue. Those parts of the side-chains deleted in the mutagenesis experiments have been coloured green and given dot surfaces to show the potential cavities left behind.
click for gif and link
to pdb.
![]() The Major Factors Affecting Protein
Stability
The Major Factors Affecting Protein
Stability![]() Hydrogen
Bonds
Hydrogen
Bonds![]() Beginning
Beginning