|
|
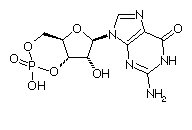
| Take a look at the figure on the right. This is the 2D diagram of a molecule, whose
significance and function we are looking for. |
 |
| |
| If your chemistry/biochemistry is dusty, you are unlikely to know
what this molecule's name is or what it does. If on the other hand you know what this is,
write down your guess, to compare it with what we are about to find. |
| |
| First, let's try to find the name of this molecule. All we have is this diagram, so
we need to look for it somewhere where we can draw the structure and use it to search for it.
The molecule looks like a nucleotide (note the nucleic-acid base, the ribose and the phosphate constituents), so it is likely to be in the database of macromolecular
structures (PDB), where many nucleotides have been co-crystallised with
proteins. To check
if we can find it there, we shall use the :
http://www.rcsb.org/ |
| |
| Clearly, we do not know the molecule name or the ligand code, but RCSB PDB
allows us to draw a structure and search for it (note that we can either search
for the full structure or do a substructure match).
Click on Search, select Advanced Search
and click on Start Advanced Search. From the "Choose a query type"
menu, select Chemical Structure (SMILES) under Chemical Components. This brings up the Marvin editor
where you can draw part of the structure to search. Try to draw the molecule as you see it, without worrying about
hydrogens at this stage.
You can undo a step and delete atoms and bonds if you make mistakes.
HINT: You can draw the substructure you want using just single bonds between carbon atoms, and when
you are done, then you can
change any single bonds to double and any non-carbon atoms to N, O or P
accordingly. You do not need to draw the whole molecule! I suggest you draw the sugar ring (that's the five-membered
ring that includes an oxygen) and the phosphate group attached to it. What you choose to draw will of course
affect the results you will get back from the search, so don't draw just a C-C bond as you will get a vast
number of hits! When you finish, press Submit Query.
If you get frustrated with drawing, you can also use the following SMILES string :
|
|
O=P2(O)OCC1OCC(O)C1O2 |
| |
|
If you draw the part of the molecule I suggested and click ok, you get back the
following string (or perhaps a different one...why?): |
|
OC1COC2COP(O)(=O)OC12 |
| |
|
The number of molecules returned depends on what substructure you searched
for and on the version of the RCSB PDB database. Using the SMILES string above I often get a different
answer depending on the year I'm running the query. |
|
You should get more than 2 answers
but 35G and PCG ought to be among them,
unless you drew a very different structure. Getting different
results than expected is all part of the fun (and not necessarily always due to the updating of the
PDB database).
As an alternative, you can search for 35G and PCG, if you wish (use them in the "Chemical ID"
search). I will assume you now have retrieved these molecules.
Examining the little gifs with the 2d
diagrams of the molecules (or just looking at the molecule names!) shows me
that the two molecules are guanosine derivatives; our mystery molecule is cyclic guanosine monophosphate.
Interestingly, close examination of the two entries (35G and PCG)
shows that the formulae are the same, as are the
SMILES strings, if you ignore the stereochemistry (@) labels. So the two molecules are stereoisomers,
i.e. they differ only in their stereochemistry. Can you find the difference
by examining the two gif diagrams?(to see a larger version of these diagrams you
need to follow the links to the individual ligands by clicking, for example, on their
3-letter code. Open the links in new windows, if you want to compare them easily).
|
| |
| The answer to the previous question is that the stereochemistry
at the phosphate atom seems to be different ( can you see the difference in
the stereo SMILES strings?)
The stereochemistry of PDB ligand atoms can be determined by the
position of their atoms in space, and so a program can automatically assign stereochemistry from
3D coordinates. How about their InChI strings? Are they different or the same?
|
| |
|
By now, you have probably seen that the molecules have the same InChI strings but different stereo SMILES.
This is confusing and it might be worth looking them up somewhere else to clarify things..
We'll try PubChem, at the National Centre for Biotechnology Information in America:
Go to:
http://pubchem.ncbi.nlm.nih.gov/search/. Do not close the RCSB window
as we will need the stereo SMILES strings to search for in PubChem. |
| |
| Go to Current System. Select the Substructure/Superstructure
tab, and then select the CID, SMILES/SMARTS,InChI tab.
Enter the stereo (isomeric) SMILES string for 35G in the box and
press the plus button next to Options to expand them. Then, check that the match stereochemistry option is set to Exact. Finally, press
Search. If the SMILES string is not recognised as being valid,
the most likely error is that you have not copied it correctly from the RCSB site, or that
the copying has introduced a space in the string (which you need to find and delete).
If the SMILES string is recognised, you will get a preview of the
substructure you put in, and this should match what you actually wanted to search for.
Keep the results(if any!) in one window and repeat the procedure for PCG.
What do you find? |
| |
|
In a previous version of PubChem both searches hit, among others, both PDB ligands.
One year the same search returned no hits for either stereo SMILES in the substructure search
(this was true even if you stripped the
hydrogens using Options. The only search that did work was the
Identity/Similarity tab. Note that you are looking for a compound
with a molecular weight of approximately 345 Da.
This year it seems that again you get no hits.
Now, use the Identity/Similarity tab and search again with the same SMILES string.
This returns compound with CID 24316, which corresponds to cyclic GMP, with the stereo SMILES string
from either 35G or PCG
- note also that there is no stereochemistry on the P atom
in the preview of the search). One may think that a substructure search should return a hit for identical structures too,
so the results of such a search could be frustrating for a user who expects the compound to be present
in the database (the whole point of this was to demonstrate how easily one can get frustrated using chemical
searching tools on the web!)
Moreover, if you check the PubChem entry (CID 24316),
you will find the PDB code 35G listed but not PCG under Names and Identifiers and,
more specifically, the Depositor Supplied Synonyms section
Until last year, this search returned both 35G and PCG.
Hence there appears to be some disagreement over whether the two PDB ligands
are actually the same or not. This is not at all unusual when it comes
to small molecule databases on the web, and when stereochemistry is
involved the probability of such problems occurring increases manyfold!
It is just one of the things that you need to be aware of...
|
| |
|
But enough with the stereochemistry problems. Let's go back to our
molecule, which we know now is cyclic GMP. This is an endogenous
metabolite in organisms, i.e. a small molecule present in the
metabolome. We still have to find out what it does. This is the subject
of the next page. |
| |


|
|


