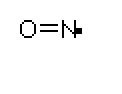|
|
|
If the molecule we are looking for is an endogenous metabolite, it will most likely
be part of the Kyoto Encyclopaedia of Genes and Genomes (KEGG). This will allow us
to find reactions in which this molecule is involved and possibly pathways that include
these reactions. Go to KEGG's Ligand database page: |
| http://www.genome.jp/kegg/ligand.html |
| |
|
Since we know the name of this molecule, we'll do a name search. In the
LIGAND Relational Database section (bottom of the page), select
Name in the Search
Compound box, type cyclic GMP
in the text box and press Go.
KEGG gets updated regularly so your answer might be slightly different but this year you will probably get:
C00942, C13817, C06194, C20640. Which one of these is our molecule?
|
| |
|
In this case you only had to search through a few molecules, and you probably found that
C00942 is our molecule, but you can imagine the problem of looking through databases using
non-standardised names, if your search had returned 200 molecules instead!
Follow the link to C00942 to see the complete entry in KEGG for this molecule.
Can you find the
name for this molecule? (hint: follow the links to Other DBs - do the other databases agree on the IUPAC name? If not, why not?).
You can also compare the InChI and InChIKeys of the same entries. What do you observe?
|
| |
|
Back to KEGG, there are two reactions listed for this molecule (what is going on
chemically in each reaction?). Note that each reaction is catalysed by two
enzymes (two distinct E.C. numbers). Two of the four E.C. numbers are phosphodiesterases
and the other two
are a type of lyase known as cyclase. According to KEGG, which ones
of these enzymes are found in humans? (hint: follow the enzyme EC number links and look at the list of genes associated with each one - HSA stands for Homo Sapiens and indicates
a human gene).
Some time ago, not all of these E.C. numbers were associated with human genes in KEGG,
but there was evidence in other databases, that all four enzymes
are found in humans. KEGG has now been updated and this discrepancy with other
databases is no longer a problem.
|
| |
|
The reactions that a metabolite takes part in represent its biochemistry. But what about
the biology related to these reactions? As any google search will tell you, one of
the most important relationships of cyclic GMP in the body is that with a tiny molecule,
nitric oxide (see diagram on the right). NO is a free radical that takes part in numerous
body functions (it is a neurotransmitter, it plays a part in the relaxation of smooth muscle,
and it functions as a hormone, among others), and it is found in the cells, in the blood and
in nervous tissue. |
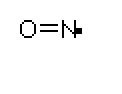 |
| One of NO's functions is to activate guanylyl cyclase and thus increase
the synthesis of cyclic GMP in various tissues (which reaction in KEGG
is the one that produces cyclic GMP?). Accumulation of cGMP leads to a reduction in
intracellular calcium and smooth muscle relaxation. Nitric oxide's regulating capabilities and its
relationship with cyclic GMP are
in fact so important that they have earned Prof. Ferid Murad a share of the Nobel prize for
Physiology/Medicine in 1998. |
| |
|
Everything that is synthesised must eventually be broken down, or it will accumulate in
the body. The reaction that breaks down cyclic GMP is catalysed by cyclic nucleotide
phosphodiesterase enzymes. But what happens if we inhibit these enzymes? Clearly, cyclic
GMP will accumulate. To find out more, let's see if we can find inhibitors for any such enzymes.
We can either search the literature, or, with a bit of luck, a structure of the enzyme
with an inhibitor may be available in the PDB. To search for such a structure go to
|
| http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/
|
|
Type in the Text search box phosphodiesterase + human + gmp
and press Search. You'll get a list of phosphodiesterases,
some of them with inhibitors. Select 1tbf and go to the
PDBsum page for it. Which phosphodiesterase is this?
Can you find using just PDBsum: a) the chemical name for the inhibitor (in this case
the ligand that resembles
cyclic GMP the most), and b) the name this inhibitor is best known by?
You can also find out a little bit more about this enzyme (e.g. in
which tissue is it expressed?) by following the link to the Protein page and
from there, the link to the UniProt entry for this protein. |
| |
|
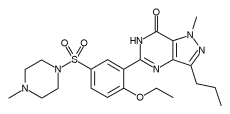
So sildenafil (see diagram on the right), better known as Viagra, is an inhibitor of PDE5,
the predominant human phosphodiesterase in the . Nitric oxide
activates the production of cGMP, which in turn accumulates and leads to smooth
muscle relaxation, and eventually penile erection. Inhibiting the enzyme that
breaks down cGMP is what leads to enhanced erections. The history of Viagra is very
interesting as it was initially designed to be a drug to treat angina. Its billion
dollar success was due, however, to the curious side effects noticed during clinical trials.
A google search will return a lot of interesting sites, should you wish to find out more.
A more scientific article with interesting facts about the drug can be found at the
Journal of the American College of Cardiology: PMID: 9935041 |
| | |
|
The specificity of sildenafil for PDE5 is what makes it a good drug. With many other
phosphodiesterases in the human body (found for example in cardiac tissue or the brain)
it is important that PDE5 be the one targeted. If, for example, PDE3 (an enzyme involved
in regulation of cardiac contractility) was seriously inhibited too, patients would
experience some serious side effects related to their heart.
This does not mean that sildenafil has no
affinity for other PDEs, just that it has much lower affinity for most of them...except one.
Apparently, the affinity of sildenafil for PDE6 (an enzyme found in the photoreceptors
of the human retina) is only approximately 10-fold lower than its affinity for its original
target, PDE5. This may be the reason why abnormalities in color vision are associated
with higher doses of Viagra. Whether the close relationship between PDE5 and PDE6 is
also responsible for the popular saying that associates blindness with a
particular sexual behaviour is anyone's guess.
|
| |
|
Inhibitors often work by mimicking the natural substrate's interactions with the target
protein. Is this the case with sildenafil? To find out, check the protein-ligand
interactions between sildenafil and human PDE5, and compare them to the interactions
between guanosine-5'-monophosphate and human PDE5, as found in the PDB structures
1t9s and 1tbf.
The quickest way to do this is to go to PDBsum:
http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ , find these two structures
and go to the ligand page for VIA (=viagra) and
5GP (=analogue of the natural substrate -
why do you think it is an analogue and not the true substrate,
cyclic GMP, that is bound in the crystal?). There
you can look at LigPlot diagrams of the most important protein-ligand interactions in the crystal.
Are there any interactions that are obviously shared by the drug and the substrate
analogue? |
| |
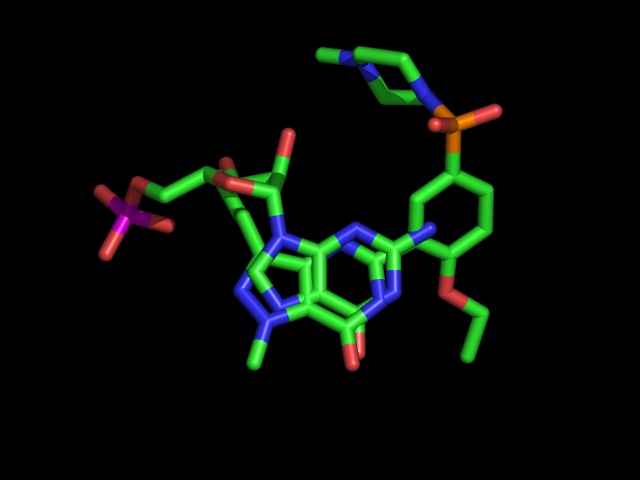
| The diagram on the right shows the relative position of the two ligands, if the
A chains of the proteins in the two crystal structures are superimposed. To get a better
look, upload both structures in Chimera, and view the ligands superimposed (
actually, the image on the left is clickable and if you have RasMol installed and your browser is properly set up
you will get a RasMol window with the superposed structures, which you can then rotate
and magnify (and whatever else you know how to do in RasMol) to get a better look. If
your browser is not setup to run RasMol scripts, just look at the static diagram next
to this text or use Chimera). Note
how nicely the two aromatic rings overlap, and how the oxygen and nitrogen pairs that
interact with Gln:A817 in both cases are aligned. As you can see from the rest of the
structures, drugs do not need to be very similar overall to their natural substrates. It
is enough if they mimic a part that has an important interaction (like the double hydrogen
bond here) that stabilises the complex. |
 |
| |
|
Finally, before we move on, let's search for any molecules that strongly resemble Viagra
and see what we can find. We shall look in PubChem, with which you are already
familiar. Go to the PubChem Compounds search:
http://pubchem.ncbi.nlm.nih.gov/search/
and choose the Structure tab and then the Similarity tab.
You will need the SMILES string for Viagra (if you have not kept it, use:
CCCc1nn(C)c2C(=O)NC(=Nc12)c3cc(ccc3OCC)[S](=O)(=O)N4CCN(C)CC4.
In the Search Options tab, select Tanimoto threshold % >= 90
(you can
choose whatever you like from the list - we do not know the distribution of similarity scores
so it is hard to tell how similar is 90% similar!).
Leave all other boxes, and press the search symbol.
|
| As the similarity score is not
very high you will get many hits (more than 1000).
You can refine your results by using the links on the right under Filters.
For example, you can select the results
that obey the
, and hence
are likely to have pharmacokinetic properties that resemble those of known drugs.
Somewhere in
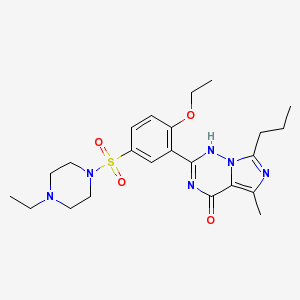
the list of this subset, you may be able to find Vardenafil (see diagram on the right), PubChem
CID=110634, another drug very
similar to Viagra which is also
used to treat erectile dysfunction. |
 |
| |
|
Now try something else yourselves: Can you find in PubChem examples
of molecules that violate the Lipinski rule of 5 but have been
tested for activity and are active against the phosphodiesterase discussed above? .
You will need to create a search that will set limits for at least one of the chemical
properties whose
ranges are explicitly mentioned in the Lipinski rule. Navigate around PubChem
to find out how to set up such a query.
(Hint: Initiate a search under the Bioactivities tab (you will need to find
the gene name for your protein which can be obtained from a link in PDBsum ) |
| |


|
|