 PPS96 Projects
PPS96 Projects  PPS96 Projects
PPS96 Projects
Cristina Cantale
The life cycle of retroviruses includes the integration of viral DNA into
cellular DNA.
Therefore retroviruses, together with retrotransposones
and transposones, are transposable elements.
Footprints of a
transposition event remain in the host DNA and often they are the only indicator
of a transposable element. At the site of an insertion, the inserted DNA is
flanked by two long terminal repeats followed by short direct repeats, whose
length is generally 5bp or 9bp. These events are repetitive and this redundance
is another signal of their presence.
Retrotransposones are closely related to retroviruses as far as the
overall organization and the ability to transpose through an RNA intermediate
are concerned. Most of them have been found as non-mobile, integrated units, but
there are others, like Tnt1 from tobacco, that are mobile. In plants, they are
described also in maize, Arabidopsis and wheat; in maize non viral
retroposons are described, too. They are also present in yeast, like Ty1, Ty3
and Ty4, in different Drosophila species, and in other insects. The main
difference with retroviruses is that they do not produce mature virus
particles, that is, they have not an extracellular phase.
They share this
aspect with transposones. In fact both of them have not life outside the genome,
they have not an independent existence and do not generate free molecules of
DNA. They range from parasites devoted to their survival to independently moving
elements involved in cellular survival and/or evolution.
Transposones are genetic elements able to move independently from
one side to an other, within and between genomes. They have DNA as genetic
material. This group is not very homogeneous, ranging from relatively simple
Insertion Elements (ISs) to complex bacteriophages. They are mainly reported in
prokaryotic organisms, but there are examples also in eukaryote (e.g. Mariner
in Drosophila). Their genomes include at least the coding regions for
those proteins involved in their own transposition, (like the insertion
sequences, identified by IS followed by a number), but they can also include
drug resistence (or other) markers. In this case they are referred to as Tn,
followed by a number.
Between more complex transposases, bacteriophage Mu
has been extensively studied and was fundamental for the understanding of the
mechanism of transposition.
Beside distinctive, specific features, there is an overall similarity between all these transponsable elements in the mechanism of transposition (or integration):
In addition, the most similar elements share the presence of a subterminal
5'CA3' nucleotide sequence.
This repetitive pattern suggests the existence
of a similarly conserved molecular mechanism to carry out the transposition.
Indeed the three steps, that is, formation of a preintegration complex,
processing at CA terminus and transfer into host DNA, have been found in all
these elements.
Sequence comparisons have been carried out either inside
the groups or among them, with the aim of confirming the similarity and
analyzing their evolutionary relationships.
When possible, the complete
genomic sequences have been compared, or large fragments of them. Alternatively,
the best conserved pol genes have been compared , eventually looking at
other viral products, as a further confirmation of conclusions. Phylogenetic
trees have been built, often containing evaluations of rates of changes.
The
relevance of horizontal transfer is a problem always present in these analyses.
It assumes a special meaning when speaking about transposable elements and their
evolutionary relationships, because of their inherent nature. Recombination has
been proposed to explain the diffusion of Mariner betweeen various, largely
different hosts and for IS3 and IS630-Tc1 families of transposones (Doak
T.G. et al., 94). The same events have been pointed out into the
integrases family, involving envelope regions and primer-binding sites (McClure
M.A. et al., 88).
Retroviruses have been extensively analyzed, especially in an attempt to
understand the origin of AIDS.
The overall conclusion which can be derived
from these studies is that lenti- and onco-viruses share a common ancestor, but
then they diverged and evoluted separately.
There is general agreement
between different authors (Sagata N et al ., 85 - Sonigo
P. et al., 85 -
Chiu I. et al., 84), about the oncoviruses phylogenetic tree,
which has been simplified in Fig. 6.
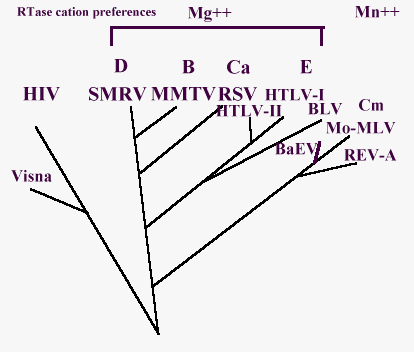
Look here for abbreviations.
A phylogenetic tree for lentiviruses has been designed by the observations about their genome by Querat and coworkers and has been summarized in this Fig. 7 (Querat G. et al., 89). There is a rough agreement about it, with different evaluations of the time of divergence (Sharp P.M. and Li W., 88 - Stephen R.M. et al., 86).
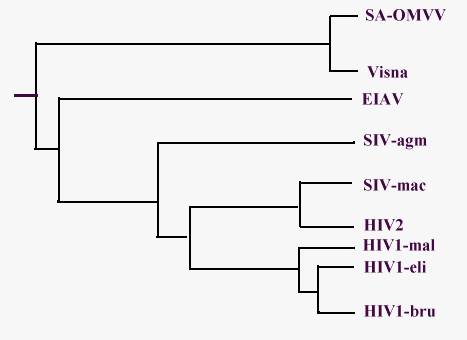
Look here for abbreviations.
Other analyses were carried out comparing protein sequences from different
sources. In this case, the analysis of evolutionary relationships is mediated
through the observation of residues and/or motifs conserved and perhaps
critical.
The central core of integrases appears to be a highly conserved
region between integrases. Apart from the DDE motif, there is a region about 50
resides long embedding them, with further identities and a high level of
conservative substitutions for about 30 residues. In addition, all the sequences
are aligned without gaps or deletions, in this region (Khan E. et al., 90
- KulkoskyJ. et al. 92 - Engelman A. and Craige R., 92).
This
is true also for transposases from the IS3 family (one of the largest group of
bacterial insertion sequences) and for a large group of other transposases from
IS6, Mariner, IS630-Tc1 families that have a less conservative signature. Thus,
it has been proposed that the majority of bacterial elements carry such a motif.(Polard
P. and Chandler M., 95)
![]() PPS96
PPS96![]() List of Contents
List of Contents![]() A bit of
history
A bit of
history![]() References
References
Last updated 25th Oct '96