 PPS96 Projects
PPS96 Projects  PPS96 Projects
PPS96 Projects
Cristina Cantale
Genome structure and life cycle
Retroviruses are a fairly homogeneus group of elements. Their genome is a
single-stranded RNA. As viral RNA itself code for proteic products, they are
referred as plus strand viruses.
Their life cycle can be summarized
in the following main steps:
These events have further consequences, besides virus replication: if
the provirus occurs in a germ cell, it remains in the host genome as an
endogenus provirus and is transmitted within the germ line. Furthermore, their
capacity of integration, transposition and complementation can produce the
conditions for cell transformation towards oncogenesis.
The retroviral
genome is generally organized in three or four different ORFs (Open Reading
Frame), called gag, pr, pol, and env, coding for different
proteic products.

Further small regions between pol and env or between env
and U3 are characteristic of different sub-families of retroviruses.
The
viral RNA has direct repeats at its ends, referred to as R in Fig.1. Both of
them are flancked by two U segments, whose name mean they are unique at 5' (U5)
and 3' (U3) ends.
Comparing genomes from different retroviruses, it appears
that
gag and even more
pol are the best conserved regions, with a slower rate of change. Env
is the most variable region. This result can be easily explained by the function
of env proteins: the envelope of the virus have to change rapidly to elude the
host immunological defense system and to spread more largely.
After
infection, the virion releases its content into the cytoplasm. As the viral RNA
looks like a conventional mRNA, capped at 5' end and polyadenylated at 3' end,
the first ORF is translated into the gag polyprotein, forming the nucleocapside
proteic portion. With different mechanisms from different viruses,
(suppression or frameshifting), the gag termination signal can be bypassed and
the mRNA translated as a gag-pol precursor; this is then proteolytically
processed to the same gag proteins, and a Reverse Transcriptase (RT,
about 90 kDa), including a ribonuclease H activity and an integrase (IN,
about 40 kDa), showing an endonuclease activity.
Due to the different
efficiency, the gag polyprotein is about 20 times more abundant than gag-pol
polyprotein.
The protease (pr) can have its own ORF or can be
included in gag-pol genes, resulting in a proteasic portion of the polyprotein
which processes itself.
The env polyprotein is expressed by a
splicing mechanism producing a shorter, subgenomic messanger, which is
translated into a polyprotein and cleaved into two proteins forming the viral
envelop.
In Fig. 1 the different proteic products are also summarized.
DNA produced by RT is a copy of the
RNA strand, with two additional sequences at the ends. A U3 segment is added at
the 5' end and viceversa a U5 segment is added at the 3' end. So each DNA end
has the same sequence: U3RU5 (direction 5'-3') and U5RU3 (direction 3'-5'). This
is called Long Terminal Repeat (LTR). Furthemore the two LTRs end with a
same sequence, consisting of a short inverted repeat.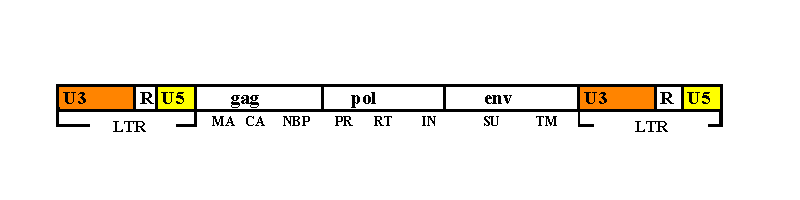
The LTRs are very important, because the very first step of integration is
represented by the interaction of the 3' end of the LTR of one of the two
strands with a IN domain. This is followed by removal of the last two
nucleotides, exposing the recessed CA(OH) 3' end which IN joins to the host
DNA. The nucleotide sequence selectivity of IN is not comparable with the
trasponsase capacity to specifically recognize its own trasponson.
In vitro studies identified only a few well-defined contacts, mainly in
the region of the terminal 5'-CA-3' dinucleotide ( this CA dinucleotide pair is
highly conserved between retroviruses) (Bushman, 1991). Probably
further features of DNA terminal end play a role into recognition, beside the
sequence.
Circular and linear forms of the viral DNA have been found after
infection, but only the linear forms appear to be integrated (Bushman,
1991 and refs).
Classification
The family of retrovirus is divided into three sub-families:
Oncoviruses are characterized by producing tumours in various vertebrate
species.
They can be transmitted as endogenous viral sequences within the
genome line and remain in an unexpressed form or behave as infective agents,
producing naturally occurring tumours.
They have been classified on the
basis of their morphological properties in four types, from A to D (Chiu
et al.84 - Sagata et al.85). Between these four different
genera, virus belonging at A, B and D genera share various aspects.
In the
following Table 1 a list of the main oncoviruses is reported.
The E type has been added later, to include morphogenic characteristic shared by BLV and HTLV-1 and 2. They have some unidentified ORF appended after env gene, not shared with other oncoviruses.
Lentiviruses show slow progressive inflammatory diseases.
Their
prototype is the Visna virus, described in 1949 for the first time, as the agent
of an epidemy among sheeps in Iceland (Sonigo et al. 85).
They
started having an important role and consequently they were extensively studied
since the discovery that the HIV virus (also referred as LAV, HTLV-III or ARV),
the ethiological agent of AIDS, is a lentiviruses.
In the following Table 2 a list of main lentiviruses is reported.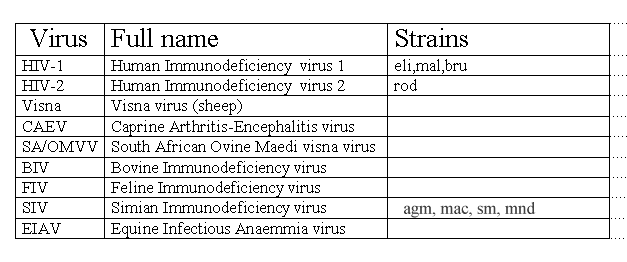
Their genome is more complex than the general type, mainly because it
contains further ORF that should be involved in regulation and control
activities.
Generally they have an ORF often referred as Q, located between
pol and env, followed by other small ORFs, one of which codes
for the tat protein, containing a conserved cysteine enriched region.
Another ORF, just before env, possibly overlapping it, codes for the
rev protein. Both of them are regulatory genes and share high genetic
variation.
Spumaviruses cause inapparent deseases in hostes and vacuolization
of cultured cells.
They are not as studied as the other sub-family, but due
to general structural similarities, a complete analogy has been assumed .
However,
Human Foam Virus (HFV) integrase, appears more closely related to
retrotransposases than to integrases, looking at multiple alignments among the
IN proteins. Moreover, a recent analysis of HFV genome (Yu et al., 96)
suggests that pol translation follows a different pathway, together with
the assembly mechanism of virions. The resulting picture should include
features of both retroviruses and hepadnaviruses, but distinct from both.
![]() PPS96
PPS96![]() List of Contents
List of Contents![]() Integrases:
a bit of history
Integrases:
a bit of history![]() References
References
Last updated 25th Oct '96