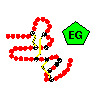 Click here for figure.
Here is the
tertiary structure.
Click here for figure.
Here is the
tertiary structure.Back to Tertiary Structure Index
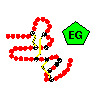 Click here for figure.
Here is the
tertiary structure.
Click here for figure.
Here is the
tertiary structure.
This results in a 'construction set' of a relatively small number of modules, from which many different proteins, called mosaic proteins, are formed, with varying lengths of polypeptide chain connecting the modules. Some proteins are formed from one or a few different modules repeated many times. There is a nomenclature of protein modules available from Peer Bork of Chris Sander's group at EMBL. Click here for a diagram of selected mosaic proteins containing the egf-like EG module, and here for those of the extracellular matrix. Illustrations of the modular nature of other types of proteins can be found in Peer Bork's list .
This phenomenon makes possible the prediction of the conformation of many polypeptides whose structures could not be deduced by other means. Once the structure of a module in isolation has been determined, for example by NMR spectroscopy, then the structure of homologous modules can be confidently predicted. Many mosaic proteins are constituents of the extracellular matrix or are membrane proteins, whose structures are difficult to determine by crystallographic methods.
The segmental nature of these proteins indicates that the different modules have had different origins during the evolution of the genome. Many modules correspond to one exon (expressed sequence in a gene; see Overview of Protein Synthesis ). It appears that mosaic proteins are the result of the duplication of exons and their shuffling between between different genes. This is more likely to occur successfully in eukaryotic cells, because of the occurrence of introns (intervening, ie unexpressed sequences), in which cleavage and splicing can occur. Mosaic proteins are particularly abundant in vertebrates. In prokaryotes, gene fusion must be precise in order to preserve the reading frame of the nucleic acid. In some bacteria the enzymes involved in trp synthesis are all encoded in different genes, whereas E. coli has two bifunctional polypeptides, each the result of the fusion of two genes.
A simple example of gene duplication occurs in the ferredoxins, where the two halves of the chain have a sequence consensus and a similar conformation. Compare the two halves of the sequence of ferredoxin in Peptococcus aerogenes:
1 10 20
A Y V I N D S C I A C G A C K P E C P V N I I Q G S
I Y A I D A D S C I D C G S C A S V C P V G A P N P E D
30 40 50
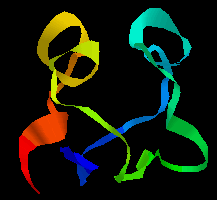 Here is the tertiary structure;the
symmetry of the two halves is apparent. Note that the C-terminal regions
of each half of the sequence would not be expected to show any homology, as
the former forms the loop between the two halves. Click
here for the structure
in RasMol (select Display:backbone or ribbons, Colours:group).
Here is the tertiary structure;the
symmetry of the two halves is apparent. Note that the C-terminal regions
of each half of the sequence would not be expected to show any homology, as
the former forms the loop between the two halves. Click
here for the structure
in RasMol (select Display:backbone or ribbons, Colours:group).
The tertiary structure of the F3 type module, first found in fibronectin, is shown below. Note that the orientation of the N- and C-terminii of the chain would allow a succession of these modules to be joined together "bead-like",as in fibronectin. This 94-residue domain is of the immunoglobulin beta-sandwich type, consisting of 7 strands forming two sheets in "Greek-key" arrangement (see future chapter on protein folds).
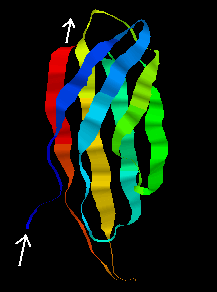 Examine the structure, which was
determined by NMR, by clicking
here.
3D images of the tertiary structures of this
fibronectin type-III module, and
other selected modules
have been prepared by Annalisa Pastore of EMBL.
Examine the structure, which was
determined by NMR, by clicking
here.
3D images of the tertiary structures of this
fibronectin type-III module, and
other selected modules
have been prepared by Annalisa Pastore of EMBL.
The average NMR structure of the F1 module from Tissue-plasminogen activator (tPA) is shown below. This is a smaller (50-residue) all-beta domain; it is involved in binding to fibrin (see below).
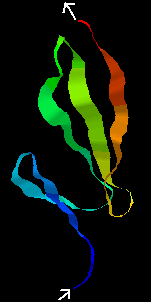 Click
The kringle fold is rich in disulphide bonds
(three are visible in the
NMR structure)
and is composed mostly of beta strands but there is one helix. This same
tPA kringle domain has also been crystallized and the structure can be seen
here (there are
3 kringle domains in the asymmetric unit).
Here is another representation of the
kringle tertiary structure.
Click
The kringle fold is rich in disulphide bonds
(three are visible in the
NMR structure)
and is composed mostly of beta strands but there is one helix. This same
tPA kringle domain has also been crystallized and the structure can be seen
here (there are
3 kringle domains in the asymmetric unit).
Here is another representation of the
kringle tertiary structure.
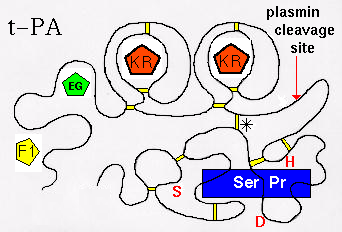
There are in fact 2 KR domains in t-PA. The tertiary structure is represented
below. (Click
here for the modular composition of t-PA, u-PA and plasminogen).
 (Diagram adapted from Kreis and Vale, 1993).
(Diagram adapted from Kreis and Vale, 1993).
The largest, C-terminal domain is the functional,
catalytic module: the serine protease (Ser Pr)domain. The function of t-PA is to
cleave a particular peptide bond in plasminogen, forming plasmin (which
is itself a serine protease). The activity
of the enzyme is markedly increased by binding to fibrin, which is effected by
the F1 domain, and the C-terminal kringle domain. Note that the serine
protease domain is connected to the
others (F1, EG, KR, KR) by a disulphide bridge (marked '*'). In fact if the
chain is cleaved at the indicated site by plasmin, the activity of the resulting 2-chain enzyme is
increased (positive feedback mechanism). t-PA is inactivated by
Plasminogen Activator Inhibitors 1 and 2 (PAI-1, PAI-2), which involves
residues Lys-296 and Arg-304 of the serine protease domain, but the C-terminal
kringle may also be involved in the initial binding of PAI-1.The residues
of the catalytic triad of the active site of the Ser Pr domain are indicated
in red (Ser,Asp,His).
The Urokinase-Type Plasminogen Activator (u-PA) functions is a similar
fashion.
Dorin-Bogdan Borza provides this material on Histidine-rich Glycoprotein (HRG).
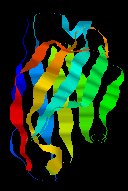 Click here
for a larger picture of the structure of the domain, which indicates the
disulphide bridge which links the two sheets of the "sandwich". See also
Annalisa Pastore's diagram of
immunoglobulin tertiary structure.
The domain may be represented like this
Click here
for a larger picture of the structure of the domain, which indicates the
disulphide bridge which links the two sheets of the "sandwich". See also
Annalisa Pastore's diagram of
immunoglobulin tertiary structure.
The domain may be represented like this 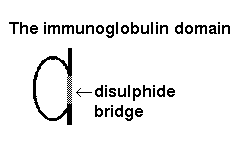
An antibody such as IgM is composed of two heavy chains, each of which consists of 4 immunoglobulin domains, and two light chains of 2 domains each.
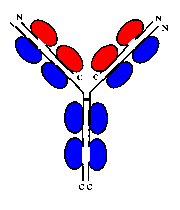 Click herefor a diagram of an antibody.
On each of the 4 chains, the N-terminal domains are known as variable domains,
as the loops connecting the beta strands are subject to exon shuffling,
creating the diversity of antibodies capable of binding to a variety of
antigens. Note however that in any one antibody molecule, the two light chains
are identical, and the two heavy chains are identical, giving two identical
antigen-binding sites at the end of each 'arm'.
Click herefor a diagram of an antibody.
On each of the 4 chains, the N-terminal domains are known as variable domains,
as the loops connecting the beta strands are subject to exon shuffling,
creating the diversity of antibodies capable of binding to a variety of
antigens. Note however that in any one antibody molecule, the two light chains
are identical, and the two heavy chains are identical, giving two identical
antigen-binding sites at the end of each 'arm'.
Papain and pepsin both cleave the heavy chains between the 2nd and 3rd domains.
Papain cleaves on the N-terminal side of the two disulphide bonds linking the two
heavy chains. This gives two separate,identical Fab fragments (The 2
C-terminal domains of the heavy chains forming an Fc fragment).
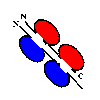 Click
here for the
crystal structure of an Fab fragment.
Click
here for the
crystal structure of an Fab fragment.
 Here is the crystal structure of a
fragment of Fab
consisting of one light chain domain and one heavy chain domain.
Here is the crystal structure of a
fragment of Fab
consisting of one light chain domain and one heavy chain domain.
Pepsin on the other hand cleaves on the C-terminal side of the disulphide linkages,
giving a single F(ab')2 fragment, consisting of 2 F(ab') fragments
(slightly longer than an Fab) linked by the two disulphide bonds (and the Fc
region is cleaved into several subfragments):
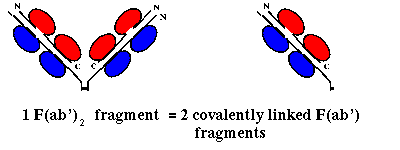 Here is the crystal structure of a
F(ab')
fragment. There are two F(ab') fragments in the asymmetric unit; this is
NOT an F(ab')2 fragment.
Here is the crystal structure of a
F(ab')
fragment. There are two F(ab') fragments in the asymmetric unit; this is
NOT an F(ab')2 fragment.
Back to Main PPS Index
J. Walshaw