Back to Protein Interactions Index

The proteins encoded by both the Cro and repressor genes bind to all three of these sites, but Cro has the highest affinity for OR3, while repressor binds most favourably to OR1. When Cro binds to OR3, RNA polymerase cannot bind to the promoter region of the repressor gene, which is therfore no longer transcribed, so that the repressor molecule is no longer produced. In the same way, with repressor protein bound to OR1, RNA polymerase is blocked from binding to the promoter of the Cro gene, and Cro synthesis ceases. Various genes involved in triggering the lytic phase lie beyond the Cro gene; their transcription is therefore also prevented by the repressor.
 Click here for a diagram
of the arrangement of the subunits in the dimer (backbone atoms only).
Click here for a diagram
of the arrangement of the subunits in the dimer (backbone atoms only).
Here is the crystal structure. There are two dimers in the asymmetric unit. Only C-alpha atoms are included.
The second and third helices of each subunit constitute the helix-turn-helix motif. The latter helix of each is termed the recognition alpha helix and binds to the DNA by insertion into the major groove. The two recognition helices of the dimer are 34Å apart, which is equivalent to one turn of the B-form of DNA. The protein binding regions (OR1, OR2 and OR3) are partly palindromic, particularly at their ends, and it is with these end regions that the recognition helices interact. The centres of the palindromic base pair sequences are separated by 10 or just over 10 base pairs depending on the binding sequence; this represents one turn of B-DNA.
 A diagram of Cro
binding to DNA.
A diagram of Cro
binding to DNA.
Click here for the pdb file (C-alpha atoms only).
The major groove of the B-DNA is widened at the ends of the binding region where the recognition helices interact, and is narrowed at the centre; the DNA is distorted. The local DNA conformation induced by the Cro is different to that which occurs when the repressor fragment (see below) is bound; this is the basis of the different affinities of Cro and repressor for the different protein binding regions.
The repressor is a dimer of a 236 amino acid subunit. Each subunit consists of two domains, one of which (the N-terminal) includes the helix-turn-helix motif and binds DNA, while the other is responsible for the formation of dimers.
However, the two domains of one subunit can be separated by mild proteolysis. Two isolated N-terminal domains can form a dimer more weakly than two complete subunits, and such a dimer has lower affinity for the DNA than the complete repressor molecule. The weakening of the dimer's binding capability by cleavage of the C-terminal domains is involved in the switching on of the lytic phase of the life cycle. When the host bacterium is exposed to ultraviolet radiation, one of its own proteins (Rec A) is converted into a proteolytic enzyme which cuts the repressor protein subunits between the two domains; the weaker DNA-binding dimer of N-terminal domains eventually disocciates from the binding sites, allowing RNA polymerase to bind to the Cro promoter to produce the Cro protein and lytic gene products. The production of new repressor molecules is prevented by Cro binding to OR3.
The N-terminal domain is topologically similar to the Cro subunit, although there are three alpha helices instead of the three beta strands.
Here is a single N-terminal domain.
In the repressor fragment (dimer of N-terminal domains), the two C-terminal helices pack against each other in a standard manner.
 Two views of the dimer of
N-terminal domains of lambda repressor
Two views of the dimer of
N-terminal domains of lambda repressor
 lambda repressor binding
to DNA
lambda repressor binding
to DNA
Click here for the pdb file.
In fact, the structure of the subunit of repressor fragment / Cro is similar to the Lambda repressor fragment, consisting of five alpha helices, although the packing arrangement of the fifth helices of each subunit against each other is different from that in Lambda repressor.
Here is the structure of a single 434 Cro subunit.
Here is the crystal structure.
Here is the structure of the 434 repressor fragment binding to DNA.
 A diagram of the CAP dimer.
This involves interaction between the long helices of each subunit.
The crystal structure of the
complete CAP dimer has been solved.
A diagram of the CAP dimer.
This involves interaction between the long helices of each subunit.
The crystal structure of the
complete CAP dimer has been solved.
 A diagram of CAP binding DNA.
This picture was obtained from a
model structure .
A diagram of CAP binding DNA.
This picture was obtained from a
model structure .
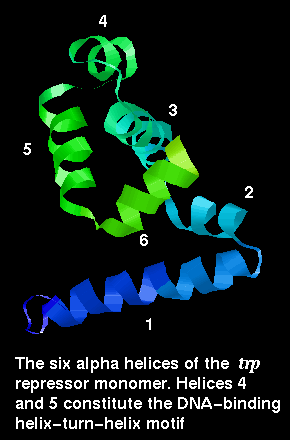
Here is the structure of a single chain from the dimer.
The arrangement of subunits in the dimer is shown below:
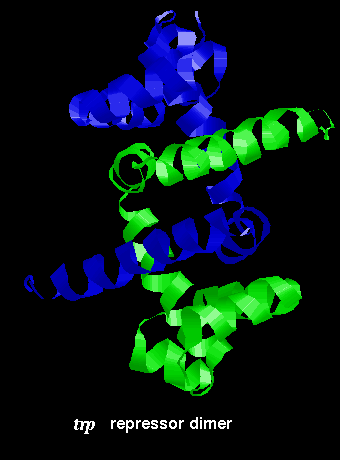
Click here for the dimer structure.
 This diagram indicates the
positions of the recognition alpha helices.
This diagram indicates the
positions of the recognition alpha helices.
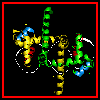 The interaction of the recognition
helices with the DNA major groove is indicated here.
The interaction of the recognition
helices with the DNA major groove is indicated here.
 A diagram
of trp repressor binding DNA from the
crystal structure.
A diagram
of trp repressor binding DNA from the
crystal structure.
The trp repressor binds to the operator region of the gene for L-tryptophan. The regulation of this gene involves negative feedback: trp repressor only binds to DNA when a L-tryptophan molecule is bound to each subunit. Each tryptophan binds between helices ....... and causes an allosteric change in the protein conformation so that its helices are in the correct orientation, 34Å apart, for interaction with the major groove. In the absence of tryptophan, the recognition helices are folded inwards towards the central core of the protein and they no longer can simultaneously fit into the groove. Therefore the repressor cannot bind, and L-tryptophan synthesis proceeds.
 This diagram indicates
the difference in conformation between the repressor (with bound tryptophan)
and the aporepressor.
This diagram indicates
the difference in conformation between the repressor (with bound tryptophan)
and the aporepressor.
Examine the structure of the aporepressor . Compare it to the repressor.
Back to Main PPS Index
J. Walshaw