|
|
Beta-strand Packing in Alpha/Beta Barrels
Packing in the core of alpha/beta barrels
The topology of alpha/beta barrels is described in Section 10. The residues of the beta-strands have a strong tendency to have hydrophobic side chains, as those on one side of the strand pack against the surrounding alpha-helices, while those on the other form the central core of the barrel.
In the classic form of the barrel, a three-residue section of each beta-strand contributes to the core. Referring to these residues as i, i+1 and i+2, alternate beta-strands have 2 (i, i+2) and 1 (i+1) side chains in the core, which therefore consists of 3 layers of 4 side chains each.
![]() This is
illustrated in the structure of glycolate oxidase,
1gox (259Kb)
[Bbk|BNL|ExP|Waw|Hal]
by this SCRIPT.
This is
illustrated in the structure of glycolate oxidase,
1gox (259Kb)
[Bbk|BNL|ExP|Waw|Hal]
by this SCRIPT.
This is also shown in this diagram (15Kb GIF), showing a top and side view.
The residues in each of the three layers are coloured blue, green and red. The orientations of each side chain are indicated. The RasMol script defines the layers as the sets top, middle and bottom, and all three collectively as core. After select-ing one of the sets, use spacefill to display the sidechains. For example to display the whole core, cut-and-paste the following:
select core spacefill select *.CA colour yellow
The result is shown in this diagram (17Kb GIF).
Alpha/beta barrels are often referred to as "TIM barrels", after Triose Phosphate Isomerase, which was the first such structure to be solved (Banner et al., 1976). However, the arrangement of side chains in the core of TIM itself is more irregular than that of glycolate oxidase. The barrel has a "height" of 5, rather than 3 residues, although the core can again be described as consisting of three layers. However, there is considerable intercalation between these layers. In the description below, which refers to the structure 1tim, each of the three is divided into two sub-layers (Figure 3).
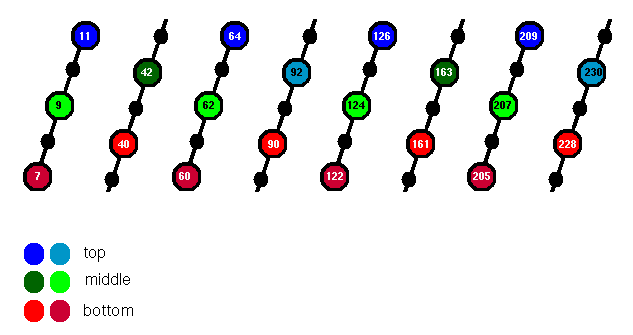
Figure 3. Five residues of each beta-strand of the TIM barrel are indicated; those whose side chains point into the core are highlighted. The top, middle and bottom layers are coloured blue, green and red respectively.
![]() Download the
triose phosphate isomerase structure, 1tim
(322Kb)
[Bbk|BNL|ExP|Waw|Hal]
and then use this RasMol SCRIPT. It defines various
sets which will be used below. The result should appear as this
diagram (18Kb GIF).
Download the
triose phosphate isomerase structure, 1tim
(322Kb)
[Bbk|BNL|ExP|Waw|Hal]
and then use this RasMol SCRIPT. It defines various
sets which will be used below. The result should appear as this
diagram (18Kb GIF).
Designating the 5 residues of each strand as i ... i+4, then alternate strands contribute the side chains of i and i+1 to the core. The two resulting sub-layers are coloured different shades of red, but it is apparent that they intercalate fairly efficiently to form a single layer, with the exception of the polar Arg 205 which is positioned underneath.
To display this layer, type:
select bottom spacefill
N.B. the sub-layers are also defined, named bottom1 and bottom2 respectively.
Alternate strands have the side chain of i+2 in the core (this sub-layer is called middle1). Two i+3 side chains (middle2) are at approximately the same level. To display the whole middle section:
select middle spacefill
The top layer is similar to the middle (inverted), consisting of two opposing i+3 side chains (top1) and the four i+4 side chains of alternate strands (top2).
select top spacefill
Note that there is considerable intercalation between the middle and top layers, so that this classification is somewhat arbitrary. The middle2 and top1 side chains are at approximately the same level. Several of the core residues are glycines, but the main chain atoms contribute.
To highlight the orientation of the side chains pointing away from the core:
select outer wireframe 40
|
|
Last updated 7th April '97