|
|
The serine proteases hydrolyze the peptide bonds of proteins. Chymotrypsin,
trypsin and elastase are the major enzymes produced by the mammalian
pancreas. They are similar in structure and function; the main chain backbone
of all 3 can be superimposed on each other very closely. The similarity of
chymotrypsin and elastase is shown below:
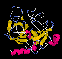 37Kb GIF
of chymotrypsin, trypsin and elastase
37Kb GIF
of chymotrypsin, trypsin and elastase
![]()
However, the sequences of the 3 proteins are only 50% homologous. Residues on the surface of the enzyme are only about 10% homologous, whereas the figure for buried residues, ie including the functionally important ones which contribute to the active site, is approximately 60%.
These three proteases are the result of divergent evolution. They differ in their specificity: chymotrypsin has a large pocket which accommodates the large hydrophobic side chains of Phenylalanine, Tyrosine and Tryptophan, and so catalyses the cleavage of peptides and esters of these amino acids. Trypsin has an Aspartate residue (189) at the bottom of the pocket (instead of Ser-189 as in chymotrypsin), and this Asp forms a salt bridge with the positively charged group at the end of the substrate Lysine and Arginine side chains, on which this enzyme acts. Elastase only accommodates small hydrophobic side chains eg Alanine, as the mouth of the pocket is partially blocked by the side chains of Val-216 and Thr-226 (these residues are both Gly in chymotrypsin).
Examine Andrew Wallace and Roman Laskowski's diagram of a tryptophan residue in the active site of chymotrypsin, created by the LIGPLOT software (Biomolecular Structure and Modelling Group, Biochemistry Department, University College, London); and the Manuel Peitsch's diagram of the enzyme highlighting the active site, in the Swiss-3DImage Collection, managed by Manuel Peitsch. There is a similar picture of elastase.
The serine proteases were so named as they have a highly reactive serine residue, Ser-195, which attacks the carboxyl group of the substrate.This results in an acylenzyme intermediate consisting of the substrate covalently bound to the enzyme at this serine.
However, the reactivity is dependent upon the arrangement of the serine side chain with two other polar side chains, approximately in a straight line, which is characteristic of all serine proteases. The Ser-195 is positioned at one end of this line, while at the other end is Asp-102, with His-57 in the middle. This is called the catalytic triad. Notice how far apart these three residues are in the sequence- the tertiary structure of the polypeptide chain brings them together in the required arrangement.
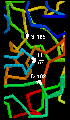 The catalytic triad is
indicated in this diagram (47Kb).
The catalytic triad is
indicated in this diagram (47Kb).
![]() You can use this SCRIPT on the
1cho structure, which displays the sidechains
of the catalytic triad, and the rest of the residues as backbone-only.
You can use this SCRIPT on the
1cho structure, which displays the sidechains
of the catalytic triad, and the rest of the residues as backbone-only.
The residues of the catalytic triad form a charge-transfer relay network. His-57, polarized by Asp-102, acts as a proton shuttle which accepts the hydrogen ion from Ser-195 as it makes a nucleophilic attack on the substrate. This mechanism will be discussed in more detail in the following pages.
The inactive precursor (the zymogen) of chymotrypsin is the 245-residue protein chymotrypsinogen.
![]() 2cga (147Kb)
[Bbk|BNL|ExP|Waw|Hal].
2cga (147Kb)
[Bbk|BNL|ExP|Waw|Hal].
A cleavage is made (by trypsin) between residues 15 and 16, to form an active form of the enzyme called pi-chymotrypsin. Further cleavages are made (by another pi-chymotrypsin molecule) to remove residues 14, 15, 147 and 148, to give the stable form of the enzyme, alpha-chymotrypsin. Note that trypsin also undergoes similar activation by means of cleavage. This is therefore a positive feedback mechanism, which activates the pancreatic enzymes in the intestine (the zymogens are secreted by the pancreatic cells).
The cleaved residues are highlighted in this SCRIPT for 2cga. The cleavage sites are displayed as thin wireframe. The thin red bond indicates the peptide bond hydrolyzed by trypsin, while the residues excised by pi-chymotrypsin are blue.
The activation of the zymogen by cleavage involves highly localized conformational changes. Cleavage between the 15th and 16th residues forms an amino-terminal group on Ile-16, which turns inwards and interacts with Asp-194 in the interior of the molecule; this stabilizes the protein. This electrostatic interaction triggers other alterations in conformation, which result in the correct arrangement of the residues forming the cavity for the substrate; this cavity is not fully formed in chymotrypsinogen.
The 1cho alpha-chymotrypsin structure is coloured by this SCRIPT to show the positions of the Ile-16 main chain (red) and the Asp-194 side chain (green), for comparison with the 2cga structure.
Differences between the structure of alpha-chymotrypsin and chymotrypsinogen
are indicated in the 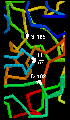 previous
diagram. Also examine Manuel Peitsch's Swiss-3DImage diagram
highlighting the different position of Ile-16 in the enzyme and zymogen.
previous
diagram. Also examine Manuel Peitsch's Swiss-3DImage diagram
highlighting the different position of Ile-16 in the enzyme and zymogen.
Some non-mammalian serine proteases have been found to have a very similar tertiary structure to their mammalian counterparts, and are 20-50% homologous with them; for example see trypsin (1sgt) from Streptomyces in a previous diagram, and also the crystal structure. However, other non-mammalian examples have no homology to the mammalian enzymes, and have completely different tertiary structure; yet the same catalytic triad, and arrangement of hydrogen bonding groups to the substrate, has evolved independently (convergent evolution). In the bacterial serine protease subtilisin, the triad consists of Ser-221, His-64 and Asp-32 as shown below:
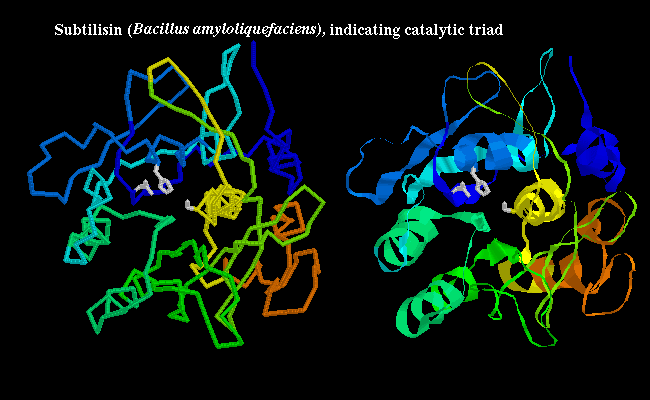
![]() subtilisin 1sbt (172Kb)
[Bbk|BNL|ExP|Waw|Hal]
...SCRIPT 1, SCRIPT 2
to highlight catalytic triad.
subtilisin 1sbt (172Kb)
[Bbk|BNL|ExP|Waw|Hal]
...SCRIPT 1, SCRIPT 2
to highlight catalytic triad.
The molecular basis of the function of chymotrypsin will be examined in more detail in the following pages.
|
|
Last updated 15th April '97