|
The DNA-binding helix-turn-helix supersecondary structure has already been
introduced (in the Supersecondary
Structure part of Section 9). A number of proteins with this motif exist
as dimers which bind to specific regions of DNA in prokaryotes, thereby
regulating transcription. The dimerization of these proteins strengthens
their interactions with DNA; the formation or dissociation of the dimers
is an intrinsic part of the regulatory mechanisms.
The lytic phase of the life cycle of certain lysogenic bacteriophages is controlled by two regulating proteins, called Cro and repressor . The genes (called Cro and repressor respectively) for these regulators are close together in the phage genome, which is integrated into the host bacterium, but are transcribed in opposite directions, with an operator region between them. On one side of this region is the promoter region for Cro, and on the other is the repressor promoter. The operator region has three protein-binding sites: OR1 , which lies inside the Cro promoter, OR3, which lies inside the repressor promoter, and OR2 , which overlaps both, as shown.
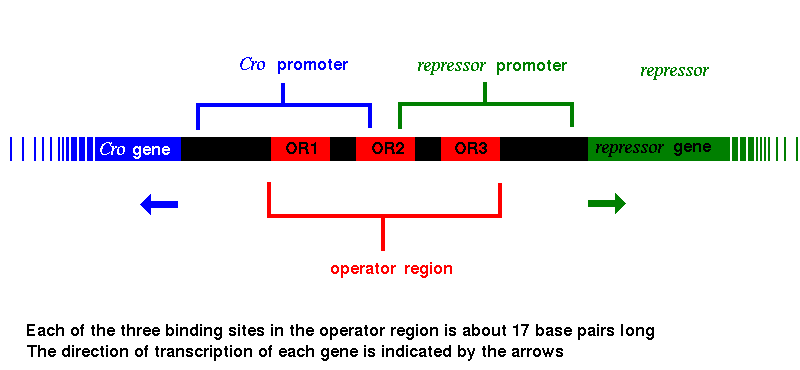
The proteins encoded by both the Cro and repressor genes bind to all three of these sites, but Cro has the highest affinity for OR3, while repressor binds most favourably to OR1. When Cro binds to OR3, RNA polymerase cannot bind to the promoter region of the repressor gene, which can therefore no longer be transcribed, so that the repressor molecule is no longer produced. In the same way, with repressor protein bound to OR1, RNA polymerase is blocked from binding to the promoter of the Cro gene, and Cro synthesis ceases. Various genes involved in triggering the lytic phase lie beyond the Cro gene; their transcription is therefore also prevented by the repressor.
The momomer of Cro from bacteriophage Lambda consists of 66 amino acids and contains three alpha helices and three beta strands. In the dimer there is an antiparallel alignment of the C-terminal strand of each subunit, giving a six-stranded antiparallel sheet.
 Click here for a diagram
of the arrangement of the subunits in the dimer (backbone atoms only).
Click here for a diagram
of the arrangement of the subunits in the dimer (backbone atoms only).
![]() Crystal structures:
Crystal structures:
The second and third helices of each subunit constitute the helix-turn-helix motif. The latter helix of each is termed the recognition alpha helix and binds to the DNA by insertion into the major groove. The two recognition helices of the dimer are 34Å apart, which is equivalent to one turn of the B-form of DNA. The protein binding regions (OR1, OR2 and OR3) are partly palindromic, particularly at their ends, and it is with these end regions that the recognition helices interact. The centres of the palindromic base pair sequences are separated by 10 or just over 10 base pairs depending on the binding sequence; this represents one turn of B-DNA.
 A diagram of Cro binding
to DNA.
A diagram of Cro binding
to DNA.
SCRIPT 2 (for structure 4cro) highlights the recognition helices in white, and displays the DNA.
The major groove of the B-DNA is widened at the ends of the binding region where the recognition helices interact, and is narrowed at the centre; the DNA is distorted. The local DNA conformation induced by Cro is different to that which occurs when the repressor fragment (see below) is bound; this is the basis of the different affinities of Cro and repressor for the different protein binding regions.
The repressor molecule of the Lambda bacteriophage binds not only to the OR1 site to prevent Cro transcription, but also to OR2, where it aids the binding of RNA polymerase to the repressor promoter. In fact a repressor dimer bound to OR1 interacts cooperatively with a second dimer binding to OR2. Therefore the "repressor " molecule acts as a gene activator as well as a repressor.
The repressor is a dimer of a 236 amino acid subunit. Each subunit consists of two domains, one of which (the N-terminal) includes the helix-turn-helix motif and binds DNA, while the other is responsible for the formation of dimers.
However, the two domains of one subunit can be separated by mild proteolysis. Two isolated N-terminal domains can form a dimer more weakly than two complete subunits, and such a dimer has lower affinity for the DNA than the complete repressor molecule. The weakening, by cleavage of the C-terminal domains, of the dimer's binding capability is involved in the switching on of the lytic phase of the life cycle. When the host bacterium is exposed to ultraviolet radiation, one of its own proteins (Rec A) is converted into a proteolytic enzyme which cuts the repressor protein subunits between the two domains; the weaker DNA-binding dimer of N-terminal domains eventually disocciates from the binding sites, allowing RNA polymerase to bind to the Cro promoter to produce the Cro protein and lytic gene products. The production of new repressor molecules is prevented by Cro binding to OR3.
The N-terminal domain is topologically similar to the Cro subunit, although there are three alpha helices instead of the three beta strands.
![]() Here is a single N-terminal domain (alpha carbons only)
1lrp (15Kb)
[Bbk|BNL|ExP|Waw|Hal].
Here is a single N-terminal domain (alpha carbons only)
1lrp (15Kb)
[Bbk|BNL|ExP|Waw|Hal].
In the repressor fragment (dimer of N-terminal domains), the two C-terminal helices pack against each other in a standard manner.
 Two views of the dimer of
N-terminal domains of lambda repressor
Two views of the dimer of
N-terminal domains of lambda repressor
 lambda repressor binding
to DNA
lambda repressor binding
to DNA
![]() PDB file 1lmb (199Kb)
[Bbk|BNL|ExP|Waw|Hal]
This SCRIPT focusses on the packing of the recognition
helix (coloured white) in the DNA major groove. Both the helices of the
DNA-binding motif are shown as space-filling.
PDB file 1lmb (199Kb)
[Bbk|BNL|ExP|Waw|Hal]
This SCRIPT focusses on the packing of the recognition
helix (coloured white) in the DNA major groove. Both the helices of the
DNA-binding motif are shown as space-filling.
In contrast to phage Lambda, in phage 434 the structures of the Cro protein (71 residues) and the DNA-binding domain (69 residues) of repressor are very similar (they have 48% sequence homology). As in Lambda, both are dimers. Again, the intact repressor molecule has a C-terminal domain to each subunit whose precise structure is not known, while the N-terminal domains can more weakly dimerize and bind to DNA.
In fact, the structure of the subunit of repressor fragment / Cro is similar to the Lambda repressor fragment, consisting of five alpha helices, although the packing arrangement of the fifth helices of each subunit against each other is different from that in Lambda repressor.
![]() a
single 434 Cro subunit 2cro (49Kb)
[Bbk|BNL|ExP|Waw|Hal].
a
single 434 Cro subunit 2cro (49Kb)
[Bbk|BNL|ExP|Waw|Hal].
![]() Here is the crystal structure 3cro (165Kb)
[Bbk|BNL|ExP|Waw|Hal].
Here is the crystal structure 3cro (165Kb)
[Bbk|BNL|ExP|Waw|Hal].
![]() N-terminal
domain of the 434 repressor subunit
1r69 (47Kb)
[Bbk|BNL|ExP|Waw|Hal]
Note the similarity to the 434 Cro structure.
N-terminal
domain of the 434 repressor subunit
1r69 (47Kb)
[Bbk|BNL|ExP|Waw|Hal]
Note the similarity to the 434 Cro structure.
![]() 434
repressor fragment binding to DNA
1per (155Kb)
[Bbk|BNL|ExP|Waw|Hal]
434
repressor fragment binding to DNA
1per (155Kb)
[Bbk|BNL|ExP|Waw|Hal]
The function of CAP is to bind to DNA to assist the binding of RNA polymerase to specific promoter regions. It is a dimer of two identical 209-residue chains. This chain has two domains: the C-terminal domain binds DNA, while the N-terminal domain (an 8-stranded jelly-roll beta barrel) is involved in interactions between the two subunits of the dimer, and also binds cyclic AMP (adenosin monophosphate). Cyclic AMP is believed to cause allosteric changes in the protein, and converts it from a non-specific DNA-binding molecule to one which binds to particular operator sites.
 A diagram of the CAP dimer.
This involves interaction between the long helices of each subunit.
A diagram of the CAP dimer.
This involves interaction between the long helices of each subunit.
![]() The crystal structure of the complete CAP dimer
3gap (303Kb)
[Bbk|BNL|ExP|Waw|Hal]
has been solved.
The crystal structure of the complete CAP dimer
3gap (303Kb)
[Bbk|BNL|ExP|Waw|Hal]
has been solved.
![]() This
picture was obtained from a model structure
2gap (52Kb)
[Bbk|BNL|ExP|Waw|Hal].
This
picture was obtained from a model structure
2gap (52Kb)
[Bbk|BNL|ExP|Waw|Hal].
The trp repressor is a DNA-binding protein which regulates tryptophan synthesis in E. coli . It is dimeric but unlike the dimers described so far, the conformation of the individual subunits is not compact, and does not occur in isolation. The chain consists of six alpha helices, including the DNA-binding helix-turn-helix motif.
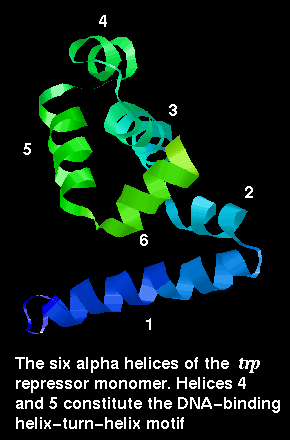
![]() Here is the structure of a single chain
1wrp (82Kb)
[Bbk|BNL|ExP|Waw|Hal]
from the dimer.
Here is the structure of a single chain
1wrp (82Kb)
[Bbk|BNL|ExP|Waw|Hal]
from the dimer.
The arrangement of subunits in the dimer is shown below:
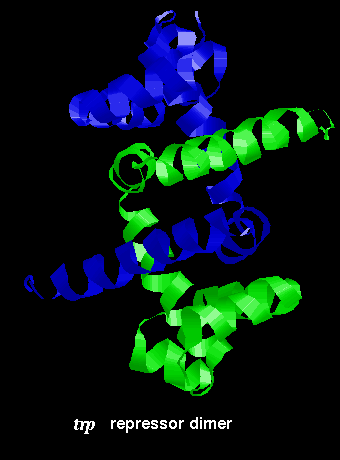
![]() Click
here for the dimer structure (155Kb). (If
you are accessing one of the PPS mirrors, i.e. not the Birkbeck College copy,
and want a chemical-MIME, rather than text, version of this file, click here
for the
original copy of the file at Birkbeck.)
Click
here for the dimer structure (155Kb). (If
you are accessing one of the PPS mirrors, i.e. not the Birkbeck College copy,
and want a chemical-MIME, rather than text, version of this file, click here
for the
original copy of the file at Birkbeck.)
This dimer was produced by applying the appropriate symmetry operation to the monomer, as detailed in REMARK 5 of the 1wrp structure.
 This diagram indicates the
positions of the recognition alpha helices.
This diagram indicates the
positions of the recognition alpha helices.
This SCRIPT renders the dimer as in the diagram.
 The interaction of the
recognition helices with the DNA major groove is indicated here.
The interaction of the
recognition helices with the DNA major groove is indicated here.
 A diagram of trp
repressor binding DNA from the crystal structure 1tro
A diagram of trp
repressor binding DNA from the crystal structure 1tro
![]() 1tro (449Kb)
[Bbk|BNL|ExP|Waw|Hal]
... SCRIPT
1tro (449Kb)
[Bbk|BNL|ExP|Waw|Hal]
... SCRIPT
The trp repressor binds to the operator region of the gene for L-tryptophan. The regulation of this gene involves negative feedback: trp repressor only binds to DNA when a L-tryptophan molecule is bound to each subunit. Each tryptophan binds between helices 3 and 5 and causes an allosteric change in the protein conformation so that its helices are in the correct orientation, 34Å apart, for interaction with the major groove. In the absence of tryptophan, the recognition helices are folded inwards towards the central core of the protein and they no longer can simultaneously fit into the groove. Therefore the repressor cannot bind, and L-tryptophan synthesis proceeds.
 This diagram indicates
the difference in conformation between the repressor (with bound tryptophan)
and the aporepressor.
This diagram indicates
the difference in conformation between the repressor (with bound tryptophan)
and the aporepressor.
![]() Examine the structure of the aporepressor
(141Kb). Compare it to the repressor. This structure
is a dimer of 3wrp (81Kb)
[Bbk|BNL|ExP|Waw|Hal],
produced by the operation described in REMARK 5
of this PDB file.
Examine the structure of the aporepressor
(141Kb). Compare it to the repressor. This structure
is a dimer of 3wrp (81Kb)
[Bbk|BNL|ExP|Waw|Hal],
produced by the operation described in REMARK 5
of this PDB file.
(If you are accessing one of the PPS mirrors, i.e. not the Birkbeck College copy, and want a chemical-MIME, rather than text, version of this file, click here for the original copy of the file at Birkbeck.)
|
Last updated 114th April '97