Many organisms are unable to synthesize some essential substances, so they must be present in these organisms' diet. Among these substances there are vitamins.
Probably the distant ancestor of human had the capability to synthesize the various vitamin as do modern plants. Today, vitamins are normally avaiable in the diets of higher organisms or are synthesize by bacteria that normally lives in their digestive system. Superfluos cellular machinery has so been lost during evolution; the protein function to bind and carry vitamins is essential.
Many of these proteins show an antiparallel beta structure. The core of these domains consists in beta strands. There can be from 4-5 to 10 strands that often build two sheets packed one against the other. Usually they can be found in a beta-barrel structure with an hydrophobic core.
Hereby we analyse the structure of two vitamin binding proteins which have a similar fold (beta-barrel) and an example of an alpha/beta domain binding vitamin B12.

Avidin, a minor constituent of egg white of reptiles, amphibia
and birds, is a glycosilated and positively charged protein while streptavidin
is non-glycosilated and neutral protein. They are secreted ad highly
stable proteins of similar size (Mr 15,800 Da per monomer) which can
bind up to four molecules of the water-soluble vitamin H, D-biotin.
Biotin is a vitamin normally involved in carboxylation-decarboxylation
reactions
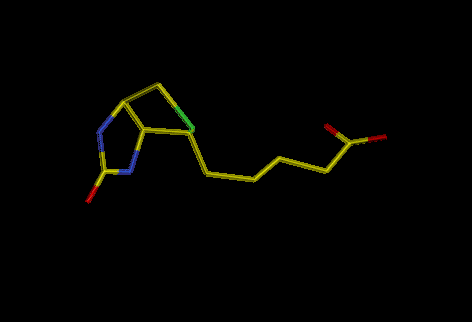
Fig. 1: biotin
The interaction with biotin is non-covalent but extremely tight, the dissociation constant of about 10E-15 M being about 1,000 to 1,000,000 times higher than that of typical antigen-antibody interactions. Their overall amino acid sequences show a low degree of similarity. Resolution of three dimensional structures of avidin (PDB file) and streptavidin (PDB file) shows them to share a high degree of structural homology. Both are tetramers of identical subunits which fold into an eight-stranded antiparallel beta-barrel. Biotin binding site within each promoter is located in a deep pocket in the core of the barrel displaying both hydrophobic and polar residues for recognition of the tightly bound vitamin and consists of residues of the barrel itself and of a loop of the adjacent subunit (Trp 110). Moreover, the binding pocket is partly closed in its outer rim by residue Trp110 of a neighboring subunit Once bound, biotin is almost completely buried in the protein core, with the exception of the valeryl side-chain carboxylate group which is exposed to solvent, hydrogen bonds to residues Ala39, Thr40 and Ser75, and triggers the formation of a network of hydrogen bonded water molecules. Two Trp residues (Trp 70 and Trp 97) and a Phe (Phe 79) are in close contact with biotin.

Fig.2: Avidin complexed with biotin

Fig.3: Biotin binding site
The biotin-protein interactions are very similar for both avidin and streptavidin.
The interset in these protein derives mainly for their pratical applications, either in vivo and in vitro, based on the rapid and almost irreverible binding to any biotin linked molecule.
In vitro they are useful for protein purification and recognition, in vivo the two proteins are used for the targeting of solid tumors.

Vitamin A (retinol) is carried by Retinol Binding Protein (RBP, PDB file), which is composed by 182 residues, from its storing site in liver to the tissue. Each RBP can transport 1 molecule of retinol; after this, it is destroyed. RBP is syntesized in the epatocytes where it binds with retinol in endoplasmic reticulum. The complex RBP - vitamin A binds to a bigger protein (prealbumin) which prevents the loss of the complex through reins. A receptor on cellular membrane let the retinol be released by the complex with a consequent structural modification of the RBP, a drastic loss of affinity for prealbumin and the secretion of "apo-RBP"

Fig.4: Vitamin A
The structure of RBP has been solved at 2 Å resolution. It shows an antiparallel eight-stranded beta-barrel. At the carbossilic end of the protein, there is an alpha-helix packed against the barrel which contains the vitamin.

Fig.5: RBP binds Vitamin A (in white)
RBP belongs to a superfamily which shows an eight-stranded beta-barrel structure and similar number of residues, all binding hydrophobic molecules. Moreover a second superfamily (P2) of small molecules exists. The proteins of this family do not show sequence homology with RBP superfamily but they have similar three-dimensional structure. On the other hand, this second family shows a beta-barrel structure built with 10 strands.

Cobalamin (Vitamin B12) and its derivatives, acts as prosthetic groups for the enzyme methionine synthase, found both in eukaryota and in prokaryotes, and for many other enzymes in prokaryotes (like glutamase mutase). These enzymes catalyze many reactions such as carbon skeleton rearrangements or the removal of a methyl group from a tertiary amine.
Methionine Syntase is a large monomeric protein composed by 1227 residues divided into two domains: the N-term domain (98 kD) contains the binding site, and the C-term domain that partecipates in reductive activation of the enzyme. The first residues at N-terminus form a helical bundle composed by two pairs of antiparallel helices. This structure reminds that of the core of the globin fold but the cobalamin lies out of the bundle while the heme is enclosed by the helices. This bundle is connected with a loop to an alpha/beta domain composed by six helices and five parallel beta strands.

Fig.6: The cobalamin-binding domains of Methionine synthase.
The alpha/beta domain of methyonine synthase shows no structural similarity to any heme binding protein although the corrin and heme macrocycles have many structural similarities.