Mutagenesis is a technique, that allows the study the structure/function relationships of proteins. Site-directed mutagenesis procedures were not 100% efficient before the advent of PCR. PCR allows the introducion of deletion, insertion and substitution mutations in DNA by simple procedures.
1. Deletions
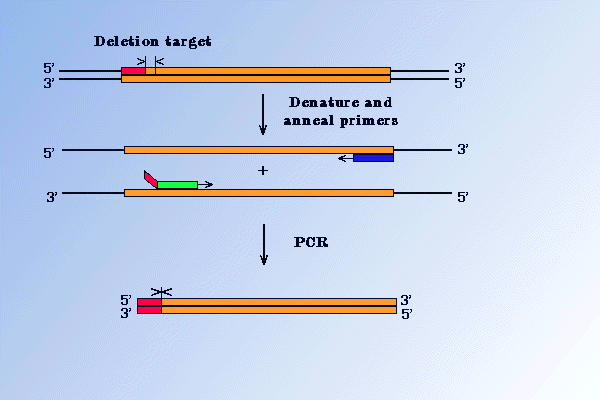
There are several methods, based on PCR, which permit the deletion of fragments from target DNA. One of these is illustrated in the Figure below. It uses a primer that spans the regions which is to be deleted. Introducing restriction enzyme sites in the mutagenic primers and in the amplified DNA, allows replacement of a wild-type sequence with mutated one by standard cloning method.

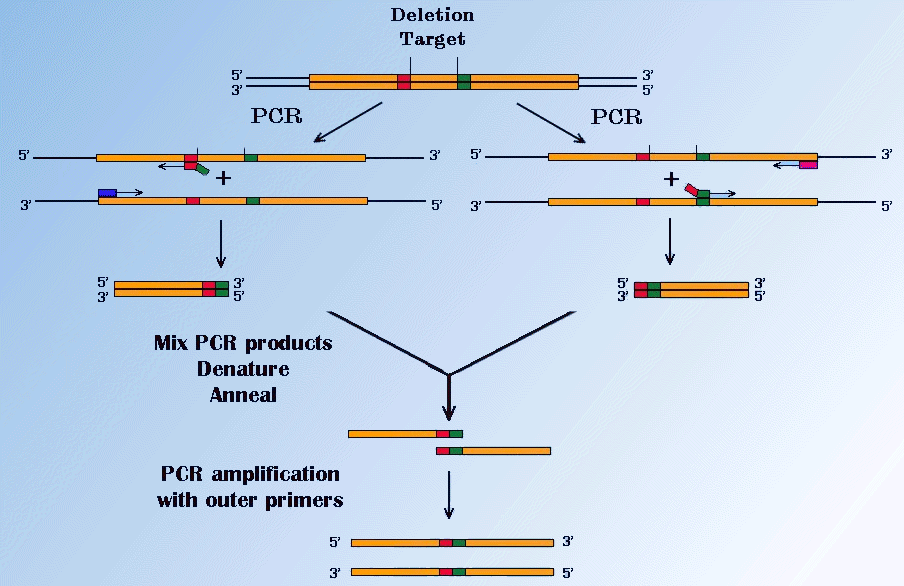
The second, popular method is the PCR - fusion. It can produce a specific deletion in a sequence at any position. Two specific fragments of the sequence are amplified in separate PCRs using primers which are complementary to each other and have 5' sequences corresponding to the fusion flanking sequence of the opposite fragment. It is illustrated in Figure below.
2. Substitutions
This is an important kind of mutation for structural biology, because it permits the changing of amino acids in proteins. There are several methods of base substitutions based on PCR. One of these in illustrated in Figure below.
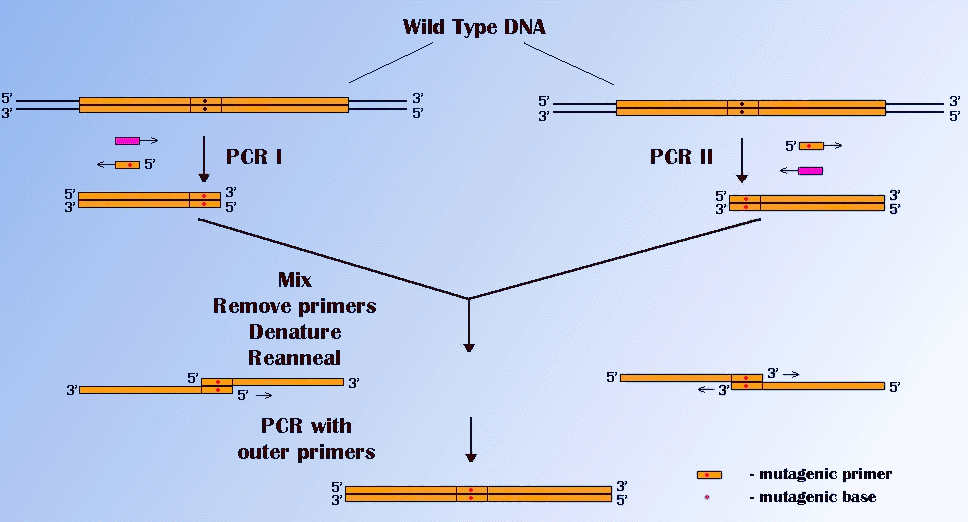
This technique uses gene fusion of fragments which have had a mutation introduced by use of mutagenic PCR primers. Two separate PCRs result in two fragments , which overlap and contain the desired mutation. They are fused and amplified in a secondary PCR.
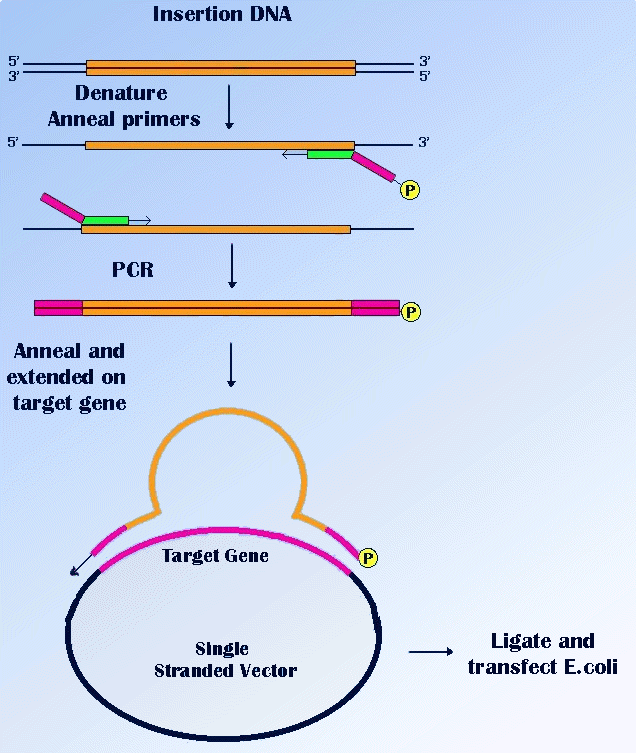
2. Insertions
The insertion of a short sequence can be performed simply, by addition of this sequence at the 5' end of a PCR primer. A suitable restriction enzyme site may also be incorporated and then the PCR product can be cloned. A method known as "sticky feet" mutagenesis, allows the introduction of large regions of DNA, into an unrelated target sequence. It is illustrated in Figure below.