
In the general search for clear pathways of predicting structure from sequence, researchers like Amos Bairoch at the Centre Medical Universitaire in Geneva and the BSM Group at University College London have been chipping away at the task by identifying significant sequence fragments which will reliably indicate a particular topology or function. Such information is valuable particularly in cases where an unknown protein does not share an overall sequence affinity with any known protein.
Such fragments generally indicate such things as:-

PROSITECreated by Amos Bairoch, this is a database of single pattern motifs. If you plug in a protein's sequence you will receive back a list of all the motifs found in that particular sequence. Searching against PROSITE can be an aid in determining the function of uncharacterised proteins translated from genomic or a cDNA sequence.
 This motif as listed in the Prosite Database consists of a 12 residue string D- x - [DNS] - {ILVFYW} - [DENSTG] - [DNQGHRK] - {GP} - [LIVMC] - [DENQSTAGC] - x(2) - [DE] - [LIVMFYW] of which the six residues
in positions 1,3,5,7,9,12 are involved in binding with Ca+ ion. In addition, this 12 residue motif is
flanked on either side with a 12 residue helix and the Ca+ is octahedrally coordinated. This motif
is found in many proteins among them actinin, aequorin, calbindin and calmodulin.
This motif as listed in the Prosite Database consists of a 12 residue string D- x - [DNS] - {ILVFYW} - [DENSTG] - [DNQGHRK] - {GP} - [LIVMC] - [DENQSTAGC] - x(2) - [DE] - [LIVMFYW] of which the six residues
in positions 1,3,5,7,9,12 are involved in binding with Ca+ ion. In addition, this 12 residue motif is
flanked on either side with a 12 residue helix and the Ca+ is octahedrally coordinated. This motif
is found in many proteins among them actinin, aequorin, calbindin and calmodulin.
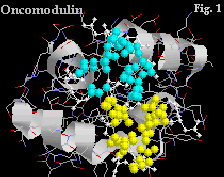
In this example of oncomodulin the atoms involved in the two Ca+ binding sites at residues 51-62 and 90-101 have been coloured yellow and blue respectively. NB the helices flanking each site.

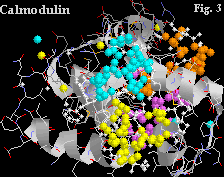
Calmodulin contains four Ca+ binding sites, shown in yellow, blue, purple and orange in Figure 2. Figure 3 shows a blown up representation of two of the sites. The similarities between the binding sites of oncomodulin and calmodulin are evident in the structural correspondences.
This database builds upon PROSITE by using its patterns, which in general correspond to the most highly conserved regions of homologous proteins, and serves as a good test for verifying sequence homology. The blocks are multiply aligned, ungapped segments corresponding to these conserved regions of proteins. Taken from the PROSITE database, the blocks are then calibrated against the SWISS-PROT database to obtain a measure of the chance distribution of matches.
PRINTS
This database is derived from OWL. Like BLOCKS, the fingerprints that it contains can encode protein folds and functionalities more flexibly and powerfully than can single motifs, therefore it serves as a complementary resource to PROSITE. PRINTS can be queried with SMITE query language which enables the user to pose complex queries.
This database analyses a protein coordinate file (PDB file) and provides details of the secondary structural motifs in the protein. (eg beta and gamma turns, beta hairpins, main-chain hydrogen bonding patterns). The database is available by anonymous ftp on 128.40.46.11. The source code can be found in the directory:- pub/promotif
PRODOM
A comprehensive collection of protein families, this database was constructed by
clustering all complete protein sequences in SWISS-PROT by the clustering algorithm Domainer
(Sonnhammer and Kahn, 1994). ProDom is different from other databases in that the modular arrangement of proteins have been taken into account and whenever domain boundaries were detected the sequences were cut to produce consistent families of
domains. The domain families produced by Domainer are stored both as multiple alignments and
consensus sequences.