 Two Dimensional NMR Spectroscopy
Two Dimensional NMR Spectroscopy Two Dimensional NMR Spectroscopy
Two Dimensional NMR Spectroscopy
Well, just have a look at this 1D NMR spectrum of a protein:
As you can see, 1D protein spectra are far too complex for interpretation as most of the signals overlap heavily. By the introduction of additional spectral dimensions these spectra are simplified and some extra information is obtained. The invention of multidimensional spectra was the major leap in NMR spectroscopy apart from the introduction of FT-NMR. Consequently, both techniques were acknowledged by a nobel prize.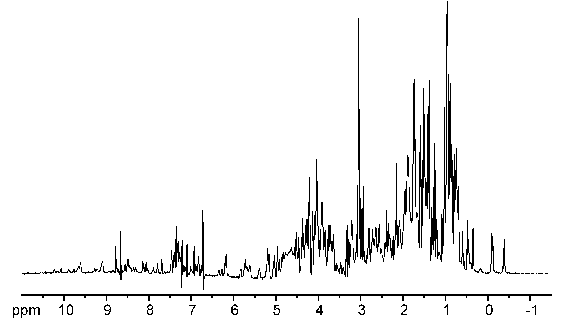
The construction of a 2D experiment is simple: In addition to preparation and detection which are already known from 1D experiments the 2D experiment has an indirect evolution time t1 and a mixing sequence. This scheme can be viewed as: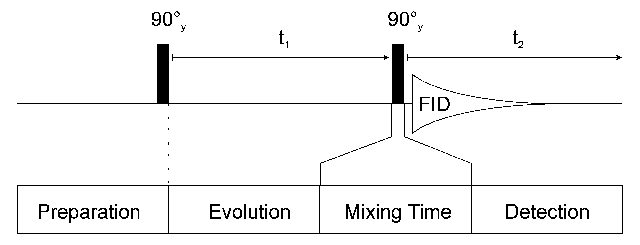
After preparation the spins can precess freely for a given time t1. During this time the magnetization is labelled with the chemical shift of the first nucleus. During the mixing time magnetization is then transferred from the first nucleus to a second one. Mixing sequences utilize two mechanisms for magnetization transfer: scalar coupling or dipolar interaction (NOE). Data are acquired at the end of the experiment (detection, often called direct evolution time); during this time the magnetization is labelled with the chemical shift of the second nucleus.
Two dimensional FT yields the 2D spectrum with two frequency axes. If the spectrum is homonuclear (signals of the same isotope (usually 1H) are detected during the two evolution periods) it has a characteristic topology:
A diagonal of signals (A and B) divides the spectrum in two equal halves. Symmetrical to this diagonal, there are more signals (X), called cross signals. The diagonal results from contributions of the magnetization that has not been changed by the mixing sequence (equal frequency in both dimensions) i.e. from contributions which remained on the same nucleus during both evolution times.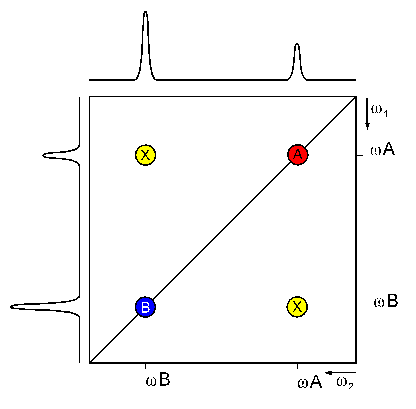
The cross signals originate from nuclei that exchanged magnetization during the mixing time (frequencies of the first and second nucleus in each dimension, respectively). They indicate an interaction of these two nuclei. Therefore, the cross signals contain the really important information of 2D NMR spectra.
In the COSY experiment, magnetization is transferred by scalar coupling. Protons that are more than three chemical bonds apart give no cross signal because the 4J coupling constants are close to 0. Therefore, only signals of protons which are two or three bonds apart are visible in a COSY spectrum (red signals). The cross signals between HN and Halpha protons are of special importance because the phi torsion angle of the protein backbone can be derived from the 3J coupling constant between them.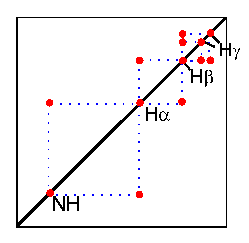
In the TOCSY experiment, magnetization is dispersed over a complete spinsystem of an amino acid by successive scalar coupling. The TOCSY experiment correlates all protons of a spin system. Therefore, not only the red signals are visible (which also appear in a COSY spectrum) but also additional signals (green) which originate from the interaction of all protons of a spin system that are not directly connected via three chemical bonds.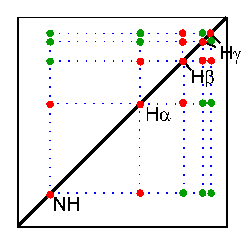
Thus a characteristic pattern of signals results for each amino acid from which the amino acid can be identified. However, some amino acids have identical spin systems and therefore identical signal patterns. They are: cysteine, aspartic acid, phenylalanine, histidine, asparagine, tryptophane and tyrosine ('AMX systems') on the one hand and glutamic acid, glutamine and methionine ('AM(PT)X systems') on the other hand.
2D NOESY:
The NOESY experiment is crucial for the
determination of protein structure.
It uses the dipolar interaction of spins (the nuclear
Overhauser effect, NOE) for correlation of protons. The
intensity of the NOE is in first approximation propotional to
1/r6, with r being the distance between the protons: The
correlation between two protons depends on the distance between
them, but normally a signal is only observed if their distance
is smaller than 5 Å. The NOESY experiment correlates all protons
which are close enough. It also correlates protons which are
distant in the amino acid sequence but close in space due to
tertiary structure. This is the most important information for
the determination of protein structures.
Here is a
picture of a 2D NOESY spectrum (38 k)
Here is a
scheme which shows the several spectral regions
in the 2D NOESY (8 k).
The most important inverse NMR experiment is the HSQC (heteronuclear single quantum correlation) the pulse sequence of which is shown above. It correlates the nitrogen atom of an NHx group with the directly attached proton. Each signal in a HSQC spectrum represents a proton that is bound to a nitrogen atom.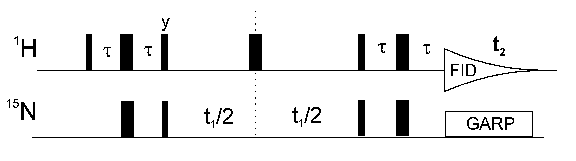
If you want to see a picture of an HSQC spectrum, here is a 10k GIF picture.
The spectrum contains the signals of the HN protons in the protein backbone. Since there is only one backbone HN per amino acid, each HSQC signal represents one single amino acid. The HSQC also contains signals from the NH2 groups of the side chains of Asn and Gln and of the aromatic HN protons of Trp and His. A HSQC has no diagonal like a homonuclear spectrum, because different nuclei are observed during t1 and t2. An analogous experiment (13C-HSQC) can be performed for 13C and 1H.