 An Overview of Terminology
An Overview of Terminology
![]() Index to Course Material
Index to Course Material
![]() Index to Section 9
Index to Section 9
These definitions are still valid, but more detail has since been added to the hierarchy. The introduction of the term "supersecondary structure" was necessary when it became clear that certain arrangements of two or three consecutive secondary structures (alpha-helices or beta-strands), are present in many different protein structures, even with completely different sequences.
Classic units of supersecondary structure include the alpha-alpha unit (two antiparallel alpha-helices joined by a 'hairpin' bend changing the chain direction by 180°); the beta-beta unit (two antiparallel strands connected by a hairpin); and the beta-alpha-beta unit (two parallel strands, separated by an alpha-helix antiparallel to them, with 2 hairpins separating the three secondary structures). Sometimes the term 'motif' is used to describe these supersecondary structures.
Other motifs of supersecondary structure have been described, such as the alpha-beta-beta and beta-beta-alpha units (Chothia, 1984).
Because supersecondary structure involves associations of secondary structures, supersecondary structure may logically be considered to be a subset of tertiary structure (see the definition above). On the other hand some literature may present supersecondary structure as a separate level in its own right.
It is important to realize that by no means all helices and strands in proteins belong to supersecondary structures. For example, proteins of the globin family consist of eight alpha-helices in contact; but the helices do not pack against other helices which are adjacent in the sequence, with the exception of the final two, which form an antiparallel helix-turn-helix motif. Again, the three- dimensional formation of these eight helices is the protein's tertiary structure.
Some relatively common combinations of the supersecondary structural motifs described above are observed in proteins. For example, there are a considerable number of proteins with a four-helix bundle, consisting of two alpha-alpha units connected by a loop. A common motif is the beta-alpha-beta-alpha-beta unit- alias the Rossman fold (effectively two consecutive beta-alpha-beta units sharing a strand). Arguably such units can be thought of as more complex supersecondary structural motifs.
These larger associations are also called (domain) folds. Some folds are considerably larger than the units described in the previous paragraph, consisting of several supersecondary structures, or secondary structures in other contexts. An example of the latter is the globin fold, six of whose helices cannot be described as a recognized supersecondary structure.
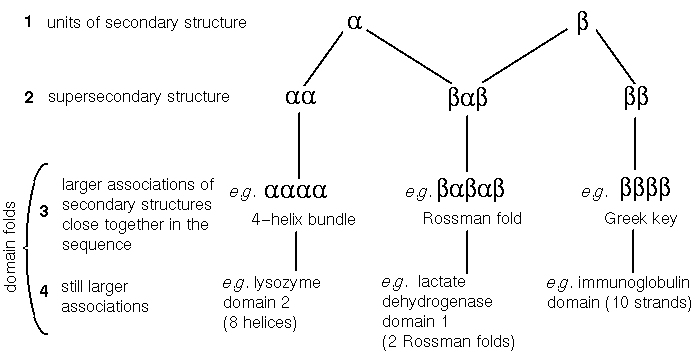
Figure 1. Levels 2, 3 and 4 come under the umbrella of 'tertiary structure', but tertiary structure can also describe how domains pack together. Possibly level 3 can be considered to constitute supersecondary structure as well as 2. Not all domain folds consist of motifs of supersecondary structure; for example the globin fold includes an alpha-alpha unit, but its remaining six helices do not comprise any 'classic' supersecondary motifs.
"Within a single subunit [polypeptide chain], contiguous portions of the polypeptide chain frequently fold into compact, local semiindependent units called domains." - Richardson, 1981Domains may be considered to be connected units which are to varying extents independent in terms of their structure, function and folding behaviour. Each domain can be described by its fold. While some proteins consist of a single domain, such as the globins described previously, others consist of several or many. A number of globular protein chains consist of two or three domains appearing as 'lobes'. In other cases the domains may be of very different nature- for example some proteins located in cell membranes have a globular intracellular or extracellular domain distinct from that which spans the membrane.
Mosaic proteins are those which consist of many repeated copies of one or a few domains, all within one polypeptide chain. Many extracellular proteins are of this nature. The domains in question are termed modules and are sometimes relatively small. Note that this term is often applied to sequences, whose structures may not be known for certain.
Tertiary structure describes the association of units within domains, but tertiary structure also includes the way in which domains fit together. This should not be confused with quaternary structure, which concerns how separate polypeptide chains associate with each other. The domain can perhaps be considered the unit of tertiary structure (c.f. helices and sheets, the units of secondary structure).
Architecture will describe the orientations of secondary structures and the way they pack together. For example, a number of folds consist of two beta-sheets packing flat against each other at 90° forming a 'sandwich'. To describe this architecture, we need not be concerned about how the strands are related sequentially in the folded chain, and in the nature of the loops which connect them; these are considerations of topology.
Note that some chains are formed by the proteolytic cleavage of an intact precursor chain. For example the active form of the digestive enzyme trypsin is formed from a single chain (the inactive form) which is cleaved into several shorter ones, without fundamentally altering the fold. Such cases should not be considered as a quaternary association of different chains.
Quaternary structure is the subject of a later chapter of the course.
Last updated 28th May '96