 Serine Proteases Part 4
Serine Proteases Part 4
![]() Index to Course Material
Index to Course Material
![]() Index to Section 12
Index to Section 12
![]() Enzymes Index;
Enzymes Index;
![]() Part 3
Part 3
![]() Part 5
Part 5
in loop
between colour in
domain strands: diagrams
______ ________ _________
Catalytic Triad His-57 side chain 1 3,4  red
Asp-102 side chain 1 5,6
Ser-195 side chain 2 3,4
Oxyanion Hole Gly-193 main chain 2 3,4
red
Asp-102 side chain 1 5,6
Ser-195 side chain 2 3,4
Oxyanion Hole Gly-193 main chain 2 3,4  white
Ser-195 main chain 2 3,4
Specificity Pocket Ser-189 side chain 2 5,6
white
Ser-195 main chain 2 3,4
Specificity Pocket Ser-189 side chain 2 5,6  green
Gly-216 side chain 2 5,6
Gly-226 side chain 2 3,4
Main Chain Ser-214 main chain 2 5,6
green
Gly-216 side chain 2 5,6
Gly-226 side chain 2 3,4
Main Chain Ser-214 main chain 2 5,6  yellow
Substrate Binding Trp-215 main chain 2 5,6
Gly-216 main chain 2 5,6
Tripeptide cleavage
yellow
Substrate Binding Trp-215 main chain 2 5,6
Gly-216 main chain 2 5,6
Tripeptide cleavage  blue
product
blue
product
The following diagrams are of chymotrypsin complexed with a tripeptide (crystal structure 8gch).
If you have not already done so, load the 8gch structure into RasMol.
![]() 8gch (185Kb) [Bbk|BNL|ExP|Waw|Hal]
8gch (185Kb) [Bbk|BNL|ExP|Waw|Hal]
SCRIPT 1 (also on the previous page) highlights the residues of interest with the colour scheme described. It defines the following RasMol sets:
These sets can be turned on and off with the commands:
select <name of set> wireframe 80 (to switch on) wireframe off (to switch off)
The tripeptide (Gly-Ala-Trp) is in fact a product of autolysis, i.e. a peptide fragment produced by the hydrolysis of chymotrypsin by another chymotrypsin molecule; refer to Part 1 . The large tryptophan side chain is situated inside the specificity pocket.
(Crystallography note: numerous different types of peptide fragment occurred in different chymotrypsin molecules in the protein crystal; this tripeptide results from the overlapping electron density of different asymmetric units.)
Views are shown both with and without the tripeptide fragment.
A view looking into the substrate-specificity pocket, without tripeptide:
If you have already applied SCRIPT 1, you can obtain a similar view by cutting and pasting the following commands:
The same view with tripeptide:
To switch on the tripeptide (chain C in the crystal structure), simply use the following commands:
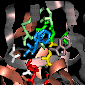 A different view to illustrate
the main-chain hydrogen bonding to the substrate/product.
A different view to illustrate
the main-chain hydrogen bonding to the substrate/product.
This view may be obtained with the commands:
To highlight the groups involved in the enzyme-substrate hydrogen bonding:
SCRIPT 3 displays, from scratch, all the residues of interest and the tripeptide highlighting enzyme-substrate binding.
Other useful renditions are those with the enzyme and product as spacefill and wireframe respectively (or vice versa); e.g. select protein and not *c. followed by spacefill.
Last updated 11th Jul '96